Regression Imputation (Stochastic vs. Deterministic & R Example)
Be careful: Flawed imputations can heavily reduce the quality of your data! Are you aware that a poor missing value imputation might destroy the correlations between your variables?
If it’s done right, regression imputation can be a good solution for this problem. I‘ll show you all the tricks you need to know in the following article.
But first of all, what is regression imputation?
Regression imputation fits a statistical model on a variable with missing values. Predictions of this regression model are used to substitute the missing values in this variable.
What are the advantages and drawbacks of regression imputation? How can I apply regression imputation? Are there even better alternatives?
Click on the buttons below to get an answer to your specific question:
Deterministic vs. Stochastic Regression Regression Imputation in R (Example) Even Better Alternatives? Ask me a Question (It's Free)
Deterministic vs. Stochastic Regression Imputation
Regression imputation consists of two subsequent steps:
- A linear regression model is estimated on the basis of observed values in the target variable Y and some explanatory variables X.
- The model is used to predict values for the missing cases in Y. Missing values of Y are then replaced on the basis of these predictions.
Relationships of X and Y (i.e. correlations, regression coefficients etc.) are preserved, since imputed values are based on regression models. This is a big advantage over simpler imputation methods such as mean imputation or zero substitution.
Regression imputation is classified into two different versions: deterministic and stochastic regression imputation.
Deterministic regression imputation replaces missing values with the exact prediction of the regression model. Random variation (i.e. an error term) around the regression slope is not considered. Imputed values are therefore often too precise and lead to an overestimation of the correlation between X and Y.
Stochastic regression imputation was developed in order to solve this issue of deterministic regression imputation. Stochastic regression imputation adds a random error term to the predicted value and is therefore able to reproduce the correlation of X and Y more appropriately.
In the following example, I’ll show you the differences between the two approaches of deterministic and stochastic regression imputation in more detail.
Regression Imputation in R (Example)
Before we can start with our regression imputation example, we need some data with missing values. So let’s create some synthetic example data with R:
# Example data set.seed(9090909) # Create reproducible data N <- 2000 # Sample size y <- round(rnorm(N, 20, 10)) # Dependent variable x1 <- round(0.2 * y + rnorm(N, 5), 2) # Predictor 1 x2 <- round(y * rpois(N, 5)) # Predictor 2 x3 <- round(0.01 * y + runif(N, 0, 10)) # Predictor 3 data <- data.frame(y, x1, x2, x3) data$y[rbinom(N, 1, 0.2) == 1] <- NA # Aproximately 10% missings in y head(data) # First 6 rows of our example data
We have created a target variable Y and three auxiliary variables X1, X2, and X3. The variable Y has some missing values, displayed as NA in rows 5 and 6. The first rows of our data set look as follows.
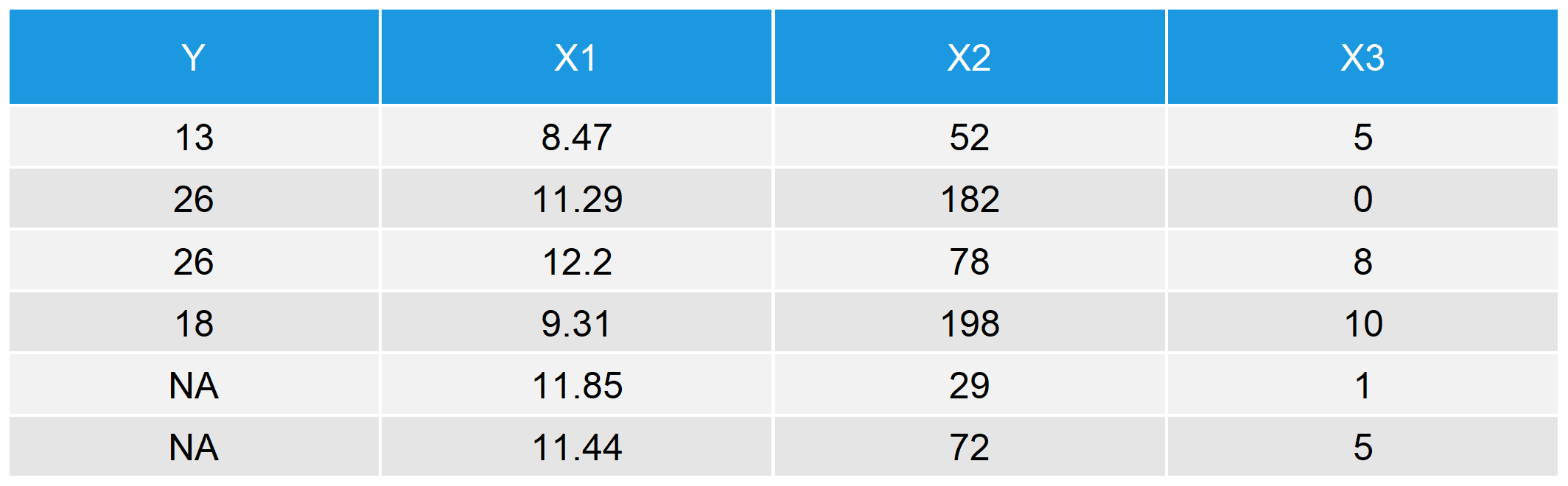
Table 1: First 6 Rows of Our Synthetic Example Data in R
A very recommendable R package for regression imputation (and also for other imputation methods) is the mice package. Install and load the package in R.
# Install and load the R package mice install.packages("mice") # Needs to be done only once library("mice") # Load package
Now, let’s apply a deterministic regression imputation to our example data. The function mice() is used to impute the data; method = “norm.predict” is the specification for deterministic regression imputation; and m = 1 specifies the number of imputed data sets (in our case single imputation).
# Deterministic regression imputation imp <- mice(data, method = "norm.predict", m = 1) # Impute data data_det <- complete(imp) # Store data
We can use almost the same code for stochastic regression imputation. We only have to change method = “norm.predict” to method = “norm.nob”.
# Stochastic regression imputation imp <- mice(data, method = "norm.nob", m = 1) # Impute data data_sto <- complete(imp) # Store data
Let’s graphically check how well our missing data imputations worked:
# Graphical comparison of deterministic and stochastic regression imputation par(mfrow = c(1, 2)) # Both plots in one graphic # Deterministic regression imputation plot(x1[!is.na(data$y)], data_det$y[!is.na(data$y)], # Plot of observed values xlim = c(0, 20), ylim = c(- 15, 60), main = "Deterministic Regression Imputation", xlab = "X1", ylab = "Y") points(x1[is.na(data$y)], data_det$y[is.na(data$y)], # Plot of missing values col = "red") abline(lm(y ~ x1, data_det), col = "#1b98e0", lwd = 1.5) # Regression slope legend("topleft", # Legend c("Observed Values", "Imputed Values", "Regression Y ~ X1"), pch = c(1, 1, NA), lty = c(NA, NA, 1), col = c("black", "red", "#1b98e0")) # Stochastic regression imputation plot(x1[!is.na(data$y)], data_sto$y[!is.na(data$y)], # Plot of observed values xlim = c(0, 20), ylim = c(- 15, 60), main = "Stochastic Regression Imputation", xlab = "X1", ylab = "Y") points(x1[is.na(data$y)], data_sto$y[is.na(data$y)], # Plot of missing values col = "red") abline(lm(y ~ x1, data_det), col = "#1b98e0", lwd = 1.5) # Regression slope legend("topleft", # Legend c("Observed Values", "Imputed Values", "Regression Y ~ X1"), pch = c(1, 1, NA), lty = c(NA, NA, 1), col = c("black", "red", "#1b98e0"))
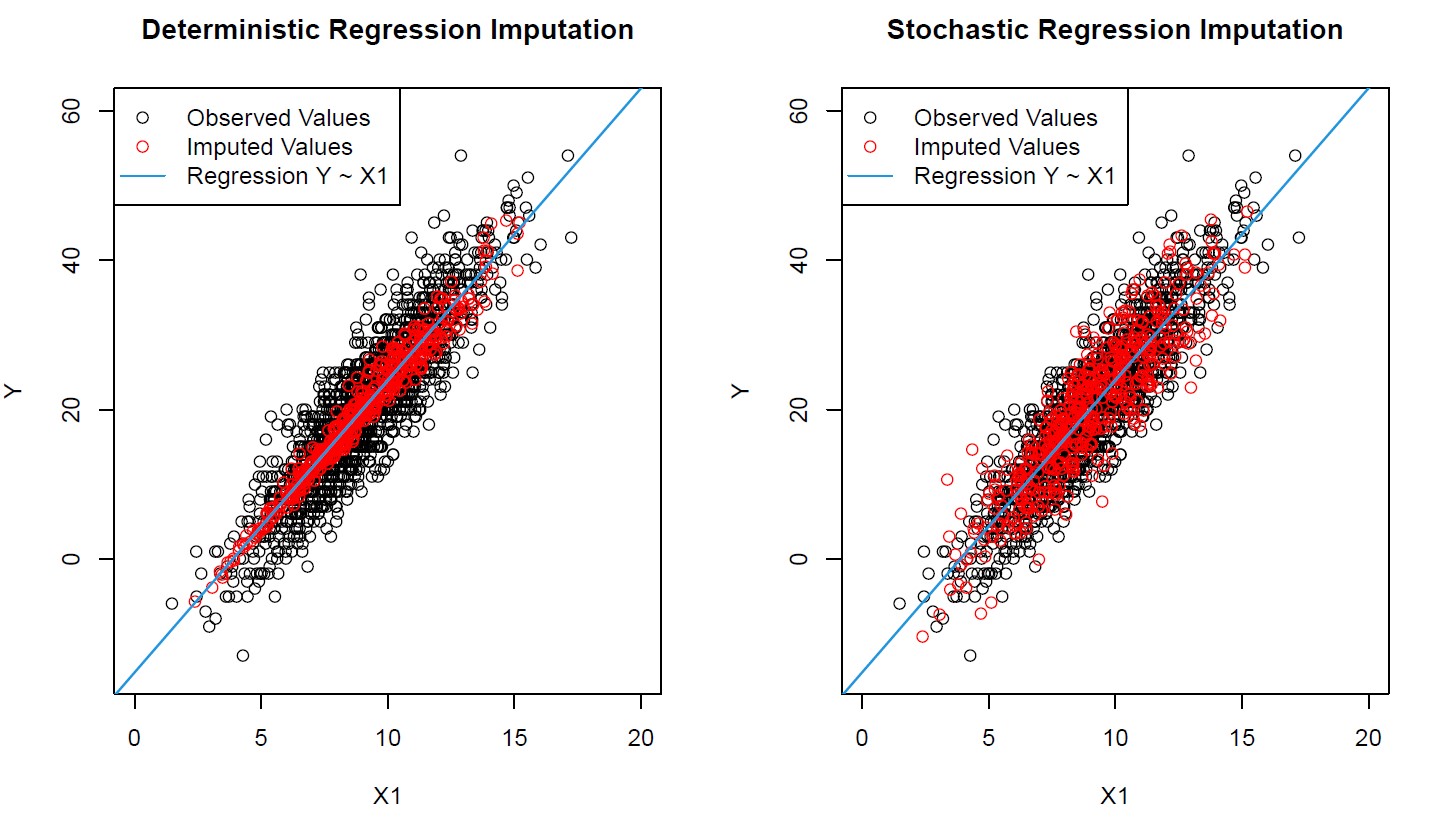
Graphic 1: Imputed Values of Deterministic & Stochastic Regression Imputation (Correlation Plots of X1 & Y)
Graphic 1 visualizes the main drawback of deterministic regression imputation: The imputed values (red bubbles) are way too close to the regression slope (blue line)!
In contrast, the imputation by stochastic regression worked much better. The distribution of imputed values is similar compared to the observed values and, hence, much more realistic.
But what does that actually mean for the analysis of our data? Have we introduced bias with the deterministic regression imputation? Let’s check some numbers:
# Correlation between X1 and Y round(cor(y, x1), 3) # True correlation ## [1] 0.897 round(cor(data_det$y, data_det$x1), 3) # Correlation after deterministic regression imputation ## [1] 0.912 round(cor(data_sto$y, data_sto$x1), 3) # Correlation after stochastic regression imputation ## [1] 0.894
The correlation coefficients between X1 and Y for our data without missings; after deterministic regression imputation; and after stochastic regression imputation confirm what we already saw graphically. While stochastic regression imputation reproduces the true correlation almost prefectly (true = 0.897; stochastic = 0.894), deterministic regression imputation overestimates the correlation (true = 0.897; deterministic = 0.912).
You might say now: OK, but does such a small difference really matter?
Answer: Yes it does. First of all, you should always avoid bias, no matter how small it is. Second, you don’t know how much bias you introduce when applying a flawed statistical method such as deterministic regression imputation. In your specific database, the bias might be much larger!
Video: Deterministic vs. Stochastic Imputation
Do you still have problems to understand the difference between deterministic and stochastic regression imputation? Don’t worry and watch the following video of Iris Eekhout’s YoutTube channel.
The video is only 17 seconds long. However, it shows perfectly the difference between deterministic regression imputation (red dots at the beginning of the video) and stochastic regression imputation (red dots at the end of the video).
Are There Even Better Alternatives? (The Drawbacks of Stochastic Regression Imputation)
At this point, we have learned that stochastic regression imputation outperforms an imputation by deterministic regression. However, the field of statistics is constantly evolving and especially one imputation method has gathered a lot of attention in recent research literature.
The method I’m talking about is called predictive mean matching (click to learn more).
But how can stochastic regression imputation be improved? Are there any drawbacks?! You guessed it, yes there are!
Drawback 1: Stochastic regression imputation may lead to implausible values. Variables are often restricted to a certain range of values (e.g. income should always be positive). Regression imputation is not able to impute according to such restrictions.
Consider the following example of implausible imputed values:
# Income data set.seed(91919) # Set seed N <- 1000 # Sample size income <- round(rnorm(N, 0, 500)) # Create some synthetic income data income[income < 0] <- income[income < 0] * (- 1) x1 <- income + rnorm(N, 1000, 1500) # Auxiliary variables x2 <- income + rnorm(N, - 5000, 2000) income[rbinom(N, 1, 0.1) == 1] <- NA # Create 10% missingness in income data_inc_miss <- data.frame(income, x1, x2) imp_inc <- mice(data_inc_miss, method = "norm.nob", m = 1) # Impute data data_inc <- complete(imp_inc) data_inc$income[data_inc$income < 0] # 10 values below 0 (unplausible) ## [1] -57.8859452 -58.9457388 -104.1099599 -147.9968026 -70.9192935 -280.2621111 ## [2] -0.5694554 -91.3508690 -76.8628876 -468.5756297
After imputing our income variable, we end up with 10 implausible values. Clearly a result we wouldn’t want to see in practice.
Drawback 2: Stochastic regression imputation leads to poor results when data is heteroscedastic. The imputation method assumes that the random error has on average the same size for all parts of the distribution, often resulting in too small or too large random error terms for the imputed values.
Consider the following example of heteroscedastic data:
# Heteroscedastic data set.seed(654654) # Set seed N <- 1:5000 # Sample size a <- 0 b <- 1 sigma2 <- N^2 eps <- rnorm(N, mean = 0, sd = sqrt(sigma2)) y <- a + b * N + eps # Heteroscedastic variable x <- 30 * N + rnorm(N[length(N)], 1000, 200) # Correlated variable y[rbinom(N[length(N)], 1, 0.3) == 1] <- NA # 30% missings data_het_miss <- data.frame(y, x) imp_het <- mice(data_het_miss, method = "norm.nob", m = 1) # Impute data data_het <- complete(imp_het) plot(x[!is.na(data_het$y)], data_het$y[!is.na(data_het$y)], # Plot of observed values main = "Stochastic Regression Imputation (Heteroscedastic Data)", xlab = "X", ylab = "Y") points(x[is.na(y)], data_het$y[is.na(y)], # Plot of missing values col = "red") abline(lm(y ~ x, data_het), col = "#1b98e0", lwd = 1.5) legend("topleft", # Legend c("Observed Values", "Imputed Values", "Regression Y ~ X"), pch = c(1, 1, NA), lty = c(NA, NA, 1), col = c("black", "red", "#1b98e0"))
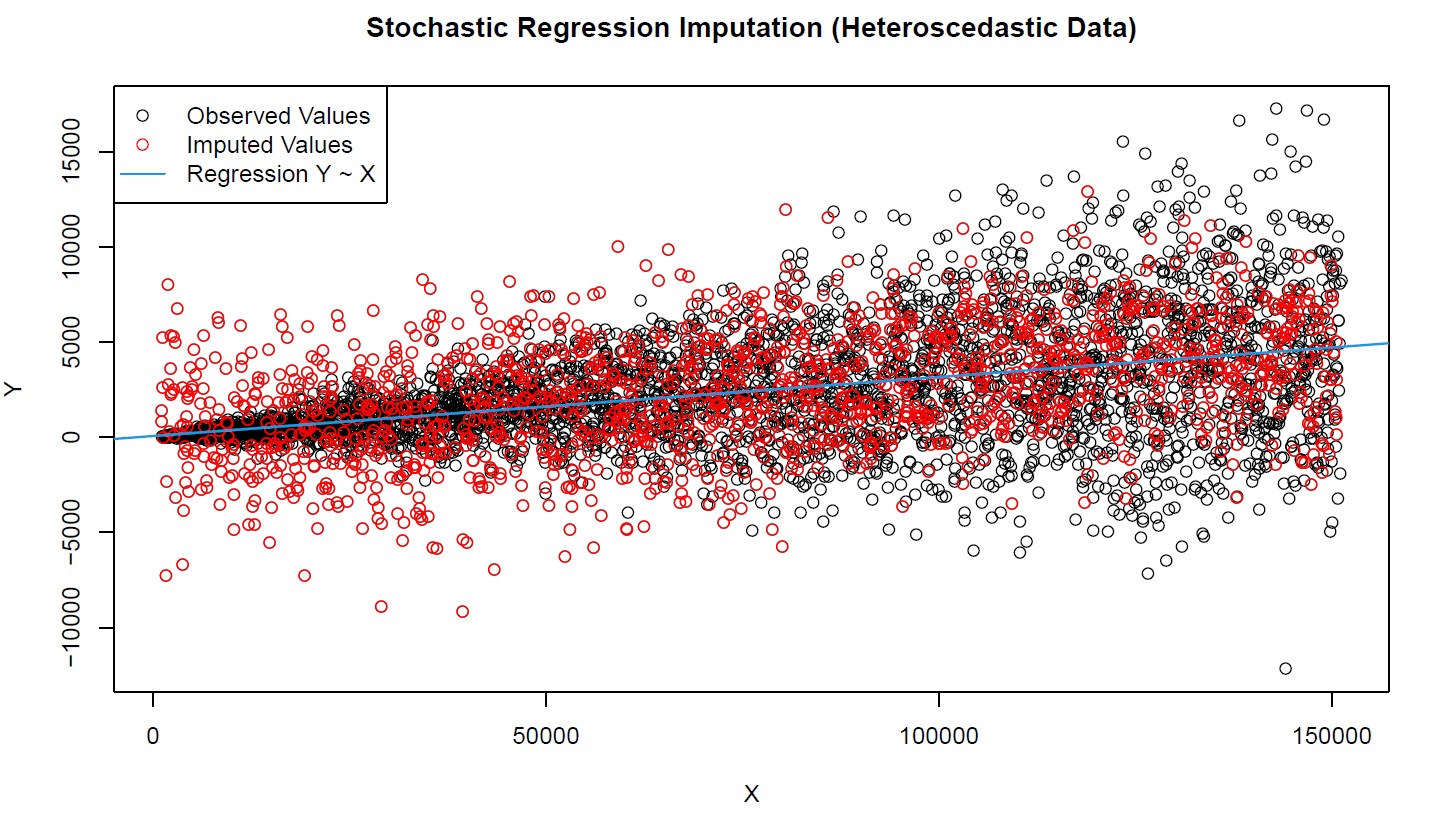
Graphic 2: Stochastic Regression Imputation of Heteroscedastic Data
Graphic 2 reveals the second major drawback of stochastic regression imputation. The observed values (black bubbles) are very heteroscedastic, i.e. the random error is much smaller for smaller values of X an Y and much larger for larger values.
This heteroscedasticity, however, is not reflected by the imputation (red bubbles). The imputed values are equally spread around the regression slope (blue line), no matter how large X and Y are. Another result we wouldn’t want to see in practice.
Find out here whether predictive mean matching is capable of solving the issues of stochastic regression imputation.
It’s Your Turn!
I showed you all the important things I know about regression imputation.
Now, I’d love to hear from your experiences! Have you already used regression imputation in practice? Have you experienced any of the drawbacks I mentioned above? Do you think about switching to newer methods such as predictive mean matching?
Let me know about your questions and experiences in the comments!
Appendix
The header graphic of this page shows a heteroscedastic relation between two variables.
Do you want to know how to create such a plot? Here’s the R code.
# Create some data n <- 1:3000 a <- 0 b <- 1 sigma2 <- n^1.9 eps <- rnorm(n, mean = 0, sd = sqrt(sigma2)) y <- - 1 * (a + b * n + eps) # Set colors palette <- colorRampPalette(colors = c("#1b98e0", "#FFFFFF")) cols <- palette(3000) par(bg = "#353436") # Produce plot plot(n, y, pch = 18, col = cols) # Estimate regression line mod <- lm(y ~ n) res <- residuals(mod) abline(coef(mod), col = "red", lwd = 3)







22 Comments. Leave new
Hi. Your post about imputation is very good, and more importantly, helpful. However, I would like to implement it in MATLAB. All resources I found online didn’t help much (maybe because I didn’t look hard enough). Is there a way I can do this?
Hi Fajrul, thank you for your kind words! Unfortunately, I am not a MATLAB user and therefore I cannot tell you, if regression imputation is available for MATLAB. However, in R the implementation is quite easy (and free). You might consider to impute your data in R and analyse the data afterwards in MATLAB.
Very interesting to read this article.I would like to thank you for the efforts you had made for writing this awesome article. This article inspired me to read more. keep it up.
Thank you Piya, that’s great to hear.
Hi, very good article. Below are few details yet that are worth mentioning:
1) Missing-data pattern that you created is so called MCAR (= missing completely at random) mechanism, e.g. line:
data$y[rbinom(N, 1, 0.2) == 1] <- NA # Aproximately 10% missings in y
This is quite unrealistic mechanism in practical data sets which are more of MAR (= missing at random) or NMAR type. For types see https://en.wikipedia.org/wiki/Missing_data#Types
Regression imputations assume are valid (provided model structure is ok) under MAR mechanism (as well as MCAR). Knowing / analyzing these mechanisms (with real data set having missingness) is very important if one wants to be able to do unbiased analysis of underlying population.
2) Single imputation provides estimates (e.g. sample mean of imputed data set) which underestimate imputation variance. This can be fixed by using multiple imputation, e.g. see https://en.wikipedia.org/wiki/Imputation_(statistics)#Multiple_imputation
3) Sampling weight adjustment (often used with incomplete survey data) can be used also to deal with missing data (= no need for imputation), e.g. see https://en.wikipedia.org/wiki/Horvitz%E2%80%93Thompson_estimator
Hi Ismo,
Thanks a lot for your positive feedback and the additional explanations. I definitely agree with all you have said, and I hope your comment helps to make these points clearer to the reader!
For more information on 1) and 2), you may also have a look here https://statisticsglobe.com/missing-data/ and here https://statisticsglobe.com/missing-data-imputation-statistics/.
Thanks!
Joachim
Thanks a lot for this!
However, I still have many missing values left after following all steps – what could be the reason?
Hey Lisa,
Thank you for the nice comment!
Could you provide the code you have used? Usually, all missing values should be gone after applying missing data imputation.
Regards
Joachim
Hi Sir:
How can find range for Intercept, Slope and R-Square?
Hey Hussein,
Are you looking for this?
https://statisticsglobe.com/extract-intercept-from-linear-regression-model-in-r
https://statisticsglobe.com/r-extract-multiple-adjusted-r-squared-from-linear-regression-model
Regards,
Joachim
This is very Interesting and productive. However, how do I handle such missing values using different techniques such as Maximum Likelihood and Expectation-Maximization techniques in R?
Hey Umar,
Thank you for the kind comment!
Please have a look at the help documentation of the mice package. The package provides numerous different imputation techniques.
Regards,
Joachim
Hi Joachim,
Your instruction on MICE is really helpful! Cheers:)
However, I am working on a longitudinal dataset. The attrition is an unavoidable issue for it. If we just simply use MICE, some TRUE missing values (e.g., death) can be incorrectly imputed. Do you have any suggestions in this case?
Thanks in advance,
Shunqi
Hey Shunqi,
Thanks a lot for the kind words! 🙂
I just came back from holidays and couldn’t reply earlier. Do you still need help with this question?
Regards,
Joachim
Hi Joachim,
No worries! I solved this problem. For those who want to deal with missing data with longitudinal data, there are many methods that may be ignored by users without carefully reading the technical document of MICE (I was!).
I attached the potential methods as follows:
method type of variable usage
2l.norm numeric Level-1 normal heteroscedastic
2l.lmer numeric Level-1 normal homoscedastic, lmer
2l.pan numeric Level-1 normal homoscedastic, pan
2l.bin binary Level-1 logistic, glmer
2lonly.mean numeric Level-2 class mean
2lonly.norm numeric Level-2 class normal
2lonly.pmm any Level-2 class predictive mean matching
Hope it helps someone!
I will definitely follow your amazing website! Keep in touch:)
All the best,
Shunqi
That’s great to hear, glad you found a solution!
Thanks a lot for sharing the additional info, and for the very kind words! 🙂
Regards,
Joachim
Do you have a good published reference on this topic?
Hey Shawn,
I think Flexible Imputation of Missing Data by Stef van Buuren is a great book when it comes to imputation techniques.
Regards,
Joachim
Thanks a lot for this article! Very helpful and easy to understand. I was wondering, what is the method to perform stochastic regression imputation in SPSS?
Hello Sophie,
I do not know SPSS, unfortunately. To my knowledge, we also do not have an author who is proficient in SPSS, unfortunately.
Regards,
Cansu
Boa tarde, estou apreciando o conteúdo desse site: Sería possivel resolver essa questão já que se trata de numeros ausentes, por favor? Eis a questão: Uma formiga encontra-se na origem de um plano cartesiano. Ela sempre se move de um ponto (X, Y) para (X+1, Y+1) ou para (X+1, Y-1). De quantas formas ela pode iniciar o movimento e chegar em (9,3), sabendo que ela passou por todos os pontos (n^2, n), com n pertencente ao conjunto dos números inteiros? Desde já obrigado.
Hello,
We mainly assist with statistical programming questions. However, you can post it on our Facebook discussion group. An interesting discussion may occur.
Best,
Cansu