R NA – What are <Not Available> Values?
Your data contains NA, <NA>, or NaN values? That’s not the end of the world — but your alarm bells should start ringing!
In R (or R Studio), NA stands for Not Available. Each cell of your data that displays NA is a missing value.
Not available values are sometimes enclosed by < and >, i.e. <NA>. That happens when the vector or column that contains the NA is a factor.
In R, NA needs to be distinguished from NaN. NaN stands for Not a Number and represents an undefined or unrepresentable value. It appears, for instance, when you try to divide by zero.
Consider the following example in R:
# Create some example variables x1 <- c(7, 9, NA, 2, 5) x2 <- as.factor(c(NA, 2, NA, 1, 1)) x3 <- c(4, NaN, 0, 9, 8) x4 <- c(6, 1, 5, 5, 7) # Create data.frame data <- data.frame(x1, x2, x3, x4)
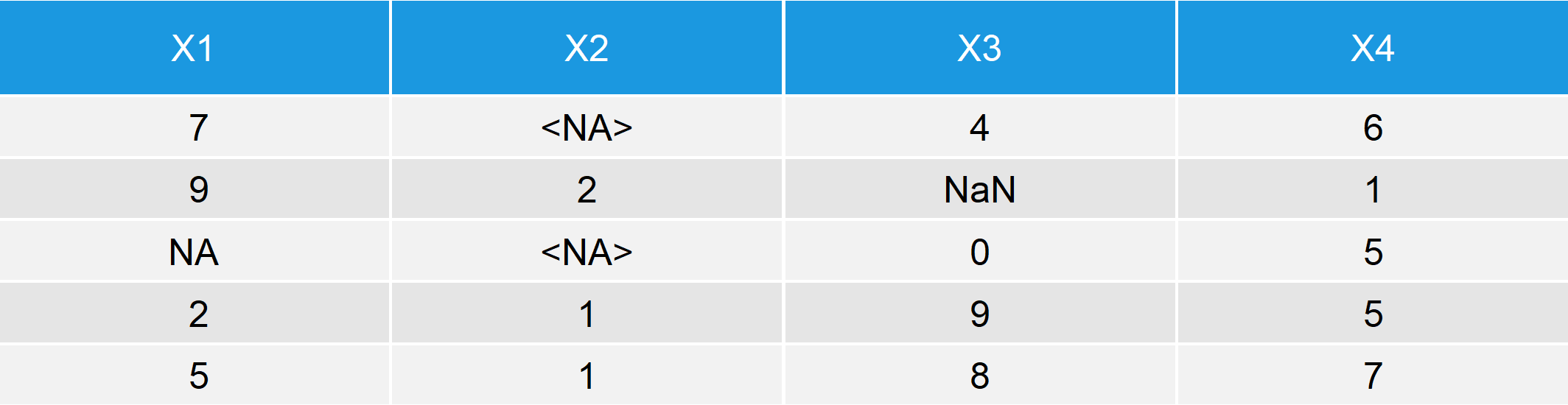
Table 1: R Example Data with NA, <NA> & NaN
The column X1 of our R example data has one missing value in the third row. The missing value is displayed with NA, since the column is numeric.
Column X2 has two missing values in the first and third row. The missings are represented by <NA>, since the second column is a factor.
The third column X3 is of class numeric (the same as X1). The second entry of the column is not a number and is therefore displayed by the code NaN.
The fourth column X4 is complete and does therefore not contain any NAs or NaNs.
Important Functions for Dealing with NAs
In the following, I’ll show you some of the most important approaches and functions of the R programming language for the handling of missing data. I’ll use our exemplifying data table that we created above.
na.omit
The na.omit function is used to exclude rows of a data set with one or more missing values. Read more…
na.omit(data) # x1 x2 x3 x4 # 2 1 9 5 # 5 1 8 7
na.omit can also be used to delete NAs in a vector…
na.omit(data$x1) # [1] 7 9 2 5
…or in a list.
# Create some data frames and matrices data_1 <- data[ , 1:2] data_2 <- data[1:3, 3:4] data_3 <- matrix(ncol = 2, c(0, NA, - 4, 3, 2, 1)) # Store data frames and matrix in list data_list <- list(data_1, data_2, data_3) # Create empty list data_list_na.omit <- list() # For loop for removal of rows with NAs in whole list for(i in 1:length(data_list)) { data_list_na.omit[[i]] <- na.omit(data_list[[i]]) }
Note: With such a for loop, all functions can be applied to a list (not only na.omit).
na.rm
na.rm is used to remove NAs of your data matrix within a function by setting na.rm = TRUE. For instance, na.rm can be used in combination with the functions mean…
mean(data$x1, na.rm = TRUE) # [1] 5.75
…and max.
max(data$x1, na.rm = TRUE) # [1] 9
use
Often confusing: The function cor uses the option use instead of na.rm.
cor(data$x1, data$x3, use = "complete.obs") # [1] -0.9011271
complete.cases
The complete.cases function creates a logical vector that indicates complete rows of our data matrix by TRUE. Read more…
complete.cases(data) # [1] FALSE FALSE FALSE TRUE TRUE
The function can also be used for casewise deletion (same as na.omit).
data[complete.cases(data), ] # x1 x2 x3 x4 # 2 1 9 5 # 5 1 8 7
is.na
is.na is also used to identify missing values via TRUE and FALSE (TRUE indicates NA). In contrast to the function complete.cases, is.na retains the dimension of our data matrix. Read more…
is.na(data) # x1 x2 x3 x4 # FALSE TRUE FALSE FALSE # FALSE FALSE TRUE FALSE # TRUE TRUE FALSE FALSE # FALSE FALSE FALSE FALSE # FALSE FALSE FALSE FALSE
!is.na
!is.na (with a ! in front) does the opposite than is.na.
!is.na(data) # x1 x2 x3 x4 # TRUE FALSE TRUE TRUE # TRUE TRUE FALSE TRUE # FALSE FALSE TRUE TRUE # TRUE TRUE TRUE TRUE # TRUE TRUE TRUE TRUE
which
Combined with the function which, logical vectors can be used to find missing values. Read more…
which(is.na(data$x1)) # [1] 3
sum
Another benefit of logical vectors is the possibility to count the amount of missing values. The function sum can be used together with is.na to count NA values in R.
sum(is.na(data$x1)) # [1] 1
summary
The summary function provides another way to count NA values in a data table, column, array, or vector.
summary(data)
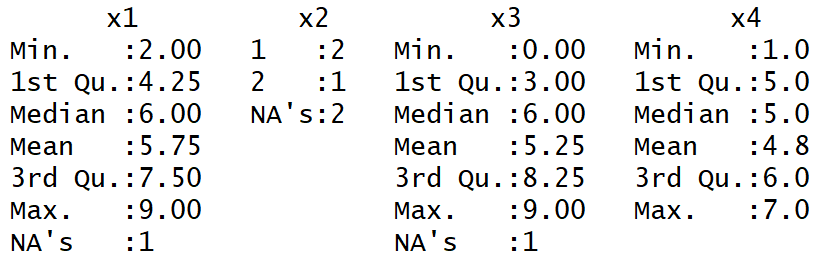
Table 2: Summary Function in R Counts NAs in Each Column
In the bottom cell of each column of Table 2, the amount of NAs is displayed.
Merge Complete Data via rbind and na.omit
The functions rbind and na.omit can be combined in order to merge (i.e. row bind) only complete rows.
# Create 2 data sets; NA in data_merge_2 data_merge_1 <- data.frame(x1 = c(5, 9, 8), x2 = c(1, 2, 3)) data_merge_2 <- data.frame(x1 = c(2, NA, 8), x2 = c(6, 9, 3)) # Merge data sets and keep only complete rows data_merge <- na.omit(rbind(data_merge_1, data_merge_2)) data_merge # Display merged data
R Remove NA, NaN, and Inf
It is also possible to exclude all rows with NA, NaN, and/or Inf values.
# Create data with NA, NaN, and Inf data_inf <- data data_inf[5, 4] <- Inf # Remove NA, NaN, and Inf data_no_na_nan_inf <- data_inf[ complete.cases(data_inf) & apply(data_inf, 1, max) != "Inf", ] data_no_na_nan_inf # Display complete subset
Recode Values to NA
Sometimes existing values have to be recoded to NA. If you want to replace a certain value with NA, you can do it as follows.
data_NA <- data # Replicate data data_NA[data_NA == 1] <- NA # Recode the value 1 to NA
If you want to recode a specific cell of your data matrix to NA, you can do it as follows.
data_NA2 <- data # Replicate data data_NA2[1, 3] <- NA # Recode row 1, column 3 to NA
Replace NAs
Logical vectors can also be used to replace NA with other values, e.g. 0. Read more…
vect_example <- data$x1 vect_example[is.na(vect_example)] <- 0 vect_example # [1] 7 9 0 2 5
Missing Value Imputation
Missing data imputation replaces missing values by new values. Data imputation has many advantages compared to the deletion of rows/columns with NAs. Read more…
In the following example, we use the predictive mean matching imputation method. However, there are many other imputation methods such as regression imputation or mean imputation available.
install.packages("mice") # Install mice package in R library("mice") # Load mice package imp <- mice(data, # Impute data m = 1, seed = 123) data_imp <- complete(imp) # Store imputed data set data_imp # Display imputed data
Video Example – How to Handle NA Values
Need more help with your NA values in R? Then you should definitely have a look at the following video of my Statistical Programming YouTube channel.
In this video, I’m explaining how to deal with incomplete data. I show easy-to-understand live examples and explain how to apply different functions such as is.na, na.omit, and na.rm.
I Would Like to Hear From You
I’ve shown you my favourite ways to handle NA values in R.
Now, I would like to hear about your experiences.
Which of these methods is your favourite? Do you use any other methods that I missed above?
Let me know in the comments!
Appendix
The header graphic of this page shows a correlation plot of two variables. Missing cases are illustrated via NA.
With the following code, the plot is created in R.
N <- 50000 # Sample size x <- rnorm(N) # X variable y <- rnorm(N) # Y variable par(bg = "#353436") # Set background color par(mar = c(0, 0, 0, 0)) # Remove space around plot plot(x, y, # Plot observed values col = "#1b98e0") points(x[1:15], y[1:15], # Plot missing values pch = 16, cex = 5, col = "#353436") text(x[1:15], y[1:15], # Write NA into each missing value "NA", col = "red")







15 Comments. Leave new
Hi Joachim,
My data has a specific column named “treatment” where the contents are 1) empty cells 2) drug 3) diet 4) unknown and 5) None.
I want to create a parallel column named “treatment_n” with drug replaced as 1 and all other content as 0.
Can you please help with this.
Thank you
Nara
Hey Nara,
that’s a great question. I have created an example, which simulates your problem. You can copy/paste the following code to your RStudio and run it yourself:
I hope that helps!
Regards,
Joachim
hi Joachim,
my data has ‘NA’ as real values (standard ISO 2 code for Namibia). How do I prevent R from seeing it as ?
Hi Adekola,
You can specify “NA” as character string or factor level. R diferentiates between “NA” and NA.
For example:
The first element is considered as country code and the second and last elements are considered as missing data.
Greetings from Germany to Namibia!
Joachim
That was great, productive and beneficial.
How about using Maximum Likelihood or Expectation-Maximization Techniques to handle the missing data?
Hey Umar,
Thank you for the kind words!
I have never done this myself, but the mlmi package seems to provide functions for Maximum Likelihood Multiple Imputation in R. Have a look here.
Regards,
Joachim
What are some possible causes of the update query in RSQLite to cause all NAs in the result set?
Date_of_Birth is an existed column but I’m trying to update it with data from another table.
DBI::dbSendQuery(con, “UPDATE DF2
SET Date_of_Birth =
(SELECT Date_of_Birth
FROM DF1
WHERE DF2.Participant_ID=DF1.Participant_ID
AND DF2.Response_ID=DF1.Response_ID)”
)
Hello,
Here are some possible causes that I found after a quick research.
Mismatched IDs: Ensure that there are matching Participant_ID and Response_ID pairs between DF2 and DF1. If no matches are found, the subquery will return NULL, which translates to NA in R.
NA Values in DF1: Check if the Date_of_Birth values in DF1 are NA for the rows where there are matching IDs. If DF1 contains NA values, these will be propagated to DF2 during the update.
SQL Syntax or Logic Error: Ensure that your SQL logic and syntax are correct. Your provided query seems fine at first glance, but there might be hidden nuances depending on your exact dataset and requirements.
Column Types: Ensure that the Date_of_Birth column in both tables are of a compatible type. If there’s a type mismatch, it might be causing unexpected behavior.
SQLite Limitations: SQLite sometimes behaves differently from other SQL-based databases. Ensure that your query is compatible with SQLite’s specific quirks and limitations.
Unintended Filtering: Ensure there’s no external or previous filtering that might be excluding certain rows from being updated.
Database Connection: Ensure your connection to the database (con in your code) is active and working correctly. Sometimes, issues can arise if the connection is unstable or has other problems.
Software or Package Bugs: While less common, there might be bugs or limitations in the RSQLite or DBI packages, or in your R version. Ensure that you’re using up-to-date versions of your software and packages.
Best,
Cansu
Thanks for that insight. The field that I’m updating has some missing values and when I used the typeof() function (dbGetQuery(mydb, “SELECT typeof(Date_of_Birth), typeof(Response_ID), typeof(Participant_ID) FROM DF2 LIMIT 1”)) to determine the data type, it was null. Is there a way to change to data type for one or two variables without creating a table schema that includes all the variables?
Please disregard duplicate above.
Thanks for that insight. The field that I’m updating has some missing values and when I used the typeof() function (dbGetQuery(mydb, “SELECT typeof(Date_of_Birth), typeof(Response_ID), typeof(Participant_ID) FROM DF2 LIMIT 1”)) to determine the data type, it was null. Is there a way to change to data type for one or two variables without creating a table schema that includes all the variables?
I’m going to try:
dbSendStatement(mydb, “UPDATE DF2 SET Date_of_Birth = cast(Date_of_Birth AS TEXT))”
Hello,
First, the SQL you provided has an error: there’s an extra closing parenthesis at the end. Secondly, before you cast and update the values, make sure to backup your table or database. Casting operations can sometimes have unintended consequences, and it’s always good to have a point to roll back to. The UPDATE statement you provided will change all Date_of_Birth values in the DF2 table to text. If there are other rows where the Date_of_Birth is already a valid date, please be aware that this would alter those values.
Alternatively, you can handle NULLs as follows.
This should only update rows in DF2 where a corresponding match is found in DF1. If no match is found, the Date_of_Birth in DF2 remains unchanged. After you run the update query, if your database system uses transactions, ensure you commit the changes. In some systems, if you don’t commit, the changes won’t be saved. After running the update, you might want to inspect some of the updated records to ensure that the data transfer occurred correctly.
I hope one of these approaches helps.
Best,
Cansu
Hi,
Thanks for the advice. I’m using the RSQLite database to operate on dataframes in R that I created from spreadsheets that came from a data extract of a clinical database of some sort. I’m going to try to set all the fields to text when I create the database tables:
dbWriteTable(mydb, “DF2”, DF2, field.types = TEXT)
And then I’ll use the query above.
Ok. I hope everything works well.
Thanks … that worked great.
#RUN vector for field.types option in dbWriteTable
(charvector1 <- rep("text",ncol(DF1)))
names(charvector1) <- names(DF1)
#change datatypes in the schema
bWriteTable(mydb, "DF1", DF1, field.types = charvector1)
Hello,
Sorry for the late response. I am glad that it finally worked! Have a good one.
Best,
Cansu