R is.na Function Example (remove, replace, count, if else, is not NA)
Well, I guess it goes without saying that NA values decrease the quality of our data.
Fortunately, the R programming language provides us with a function that helps us to deal with such missing data: the is.na function.
In the following article, I’m going to explain what the function does and how the function can be applied in practice.
Let’s dive in…
The is.na Function in R (Basics)
Before we can start, let’s create some example data in R (or R Studio).
set.seed(11991) # Set seed N <- 1000 # Sample size x_num <- round(rnorm(N, 0, 5)) # Numeric x_fac <- as.factor(round(runif(N, 0, 3))) # Factor x_cha <- sample(letters, N, replace = TRUE) # Character x_num[rbinom(N, 1, 0.2) == 1] <- NA # 20% missings x_fac[rbinom(N, 1, 0.3) == 1] <- NA # 30% missings x_cha[rbinom(N, 1, 0.05) == 1] <- NA # 5% missings data <- data.frame(x_num, x_fac, x_cha, # Create data frame stringsAsFactors = FALSE) head(data) # First rows of data
Our data consists of three columns, each of them with a different class: numeric, factor, and character. This is how the first six lines of our data look like:
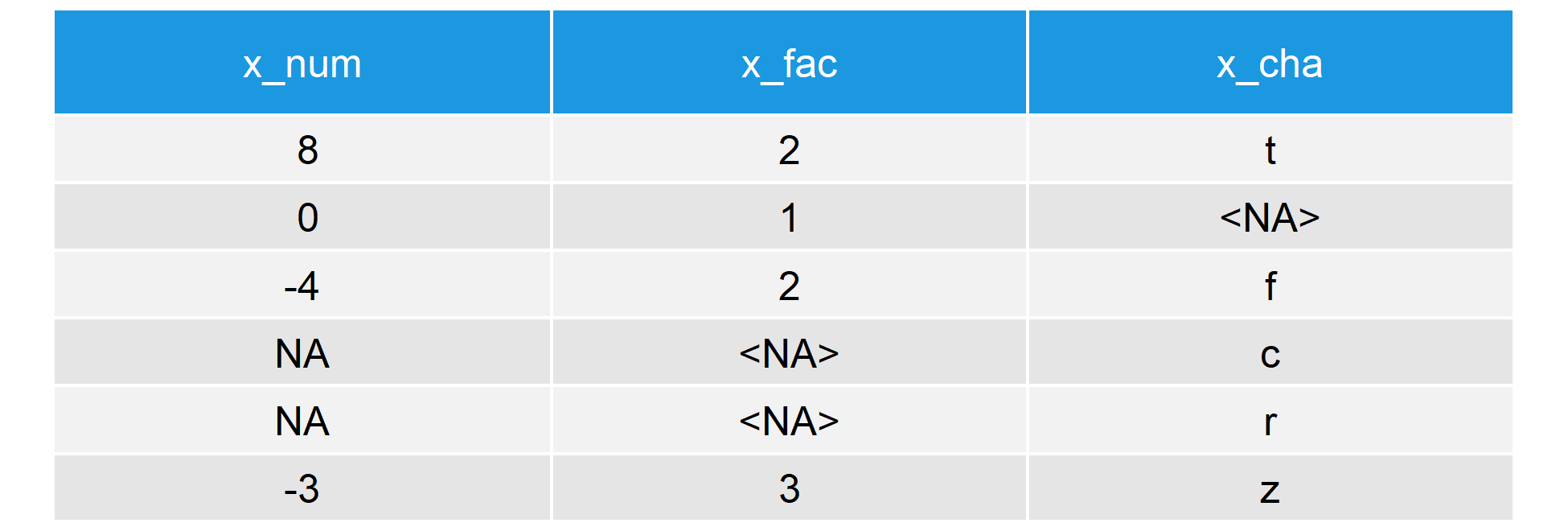
Table 1: Example Data for the is.na R Function (First 6 Rows)
Let’s apply the is.na function to our whole data set:
is.na(data) # x_num x_fac x_cha # [1,] FALSE FALSE FALSE # [2,] FALSE FALSE TRUE # [3,] FALSE FALSE FALSE # [4,] TRUE TRUE FALSE # [5,] TRUE TRUE FALSE # [6,] FALSE FALSE FALSE # ...
The function produces a matrix, consisting of logical values (i.e. TRUE or FALSE), whereby TRUE indicates a missing value. Compare the output with the data table above — The TRUE values are at the same position as before the NA elements.
An important feature of is.na is that the function can be reversed by simply putting a ! (exclamation mark) in front. In this case, TRUE indicates a value that is not NA in R:
!is.na(data) # x_num x_fac x_cha # [1,] TRUE TRUE TRUE # [2,] TRUE TRUE FALSE # [3,] TRUE TRUE TRUE # [4,] FALSE FALSE TRUE # [5,] FALSE FALSE TRUE # [6,] TRUE TRUE TRUE # ...
Exactly the opposite output as before!
We are also able to check whether there is or is not an NA value in a column or vector:
is.na(data$x_num) # Works for numeric ... is.na(data$x_fac) # ... factor ... is.na(data$x_cha) # ... and character !is.na(data$x_num) # The explanation mark still works !is.na(data$x_fac) !is.na(data$x_cha)
As you have seen, is.na provides us with logical values that show us whether a value is NA or not. We can apply the function to a whole database or to a column (no matter which class the vector has).
That’s nice, but the real power of is.na becomes visible in combination with other functions — And that’s exactly what I’m going to show you now.
R provides several other is.xxx functions that are very similar to is.na (e.g. is.nan, is.null, or is.finite). Stay tuned — All you learn here can be applied to many different programming scenarios!
is.na in Combination with Other R Functions
In the following, I have prepared examples for the most important R functions that can be combined with is.na.
Remove NAs of Vector or Column
In a vector or column, NA values can be removed as follows:
is.na_remove <- data$x_num[!is.na(data$x_num)]
Note: Our new vector is.na_remove is shorter in comparison to the original column data$x_num, since we use a filter that deletes all missing values.
You can learn more about the removal of NA values from a vector here…
If you want to drop rows with missing values of a data frame (i.e. of multiple columns), the complete.cases function is preferable. Learn more…
Replace NAs with Other Values
Based on is.na, it is possible to replace NAs with other values such as zero…
is.na_replace_0 <- data$x_num # Duplicate first column is.na_replace_0[is.na(is.na_replace_0)] <- 0 # Replace by 0
…or the mean.
is.na_replace_mean <- data$x_num # Duplicate first column x_num_mean <- mean(is.na_replace_mean, na.rm = TRUE) # Calculate mean is.na_replace_mean[is.na(is.na_replace_mean)] <- x_num_mean # Replace by mean
In case of characters or factors, it is also possible in R to set NA to blank:
is.na_blank_cha <- data$x_cha # Duplicate character column is.na_blank_cha[is.na(is.na_blank_cha)] <- "" # Class character to blank is.na_blank_fac <- data$x_fac # Duplicate factor column is.na_blank_fac <- as.character(is.na_blank_fac) # Convert temporarily to character is.na_blank_fac[is.na(is.na_blank_fac)] <- "" # Class character to blank is.na_blank_fac <- as.factor(is.na_blank_fac) # Recode back to factor
Count NAs via sum & colSums
Combined with the R function sum, we can count the amount of NAs in our columns. According to our previous data generation, it should be approximately 20% in x_num, 30% in x_fac, and 5% in x_cha.
sum(is.na(data$x_num)) # 213 missings in the first column sum(is.na(data$x_fac)) # 322 missings in the second column sum(is.na(data$x_cha)) # 47 missings in the third column
If we want to count NAs in multiple columns at the same time, we can use the function colSums:
colSums(is.na(data)) # x_num x_fac x_cha # 213 322 47
Detect if there are any NAs
We can also test, if there is at least 1 missing value in a column of our data. As we already know, it is TRUE that our columns have NAs.
any(is.na(data$x_num)) # [1] TRUE
Locate NAs via which
In combination with the which function, is.na can be used to identify the positioning of NAs:
which(is.na(data$x_num)) # [1] 4 5 14 17 22 23...
Our first column has missing values at the positions 4, 5, 14, 17, 22, 23 and so forth.
if & ifelse
Missing values have to be considered in our programming routines, e.g. within the if statement or within for loops.
In the following example, I’m printing “Damn, it’s NA” to the R Studio console whenever a missing occurs; and “Wow, that’s awesome” in case of an observed value.
for(i in 1:length(data$x_num)) { if(is.na(data$x_num[i])) { print("Damn, it's NA") } else { print("Wow, that's awesome") } } # [1] "Wow, that's awesome" # [1] "Wow, that's awesome" # [1] "Wow, that's awesome" # [1] "Damn, it's NA" # [1] "Damn, it's NA" # [1] "Wow, that's awesome" # ...
Note: Within the if statement we use is na instead of equal to — the approach we would usually use in case of observed values (e.g. if(x[i] == 5)).
Even easier to apply: the ifelse function.
ifelse(is.na(data$x_num), "Damn, it's NA", "Wow, that's awesome") # [1] "Wow, that's awesome" "Wow, that's awesome" "Wow, that's awesome" "Damn, it's NA" # [5] "Damn, it's NA" "Wow, that's awesome" ...
Video & Further Examples for the Handling of NAs in R
Do you need further info on the R code of this article? Then you might have a look at the following video on my YouTube channel. In the video, I’m explaining the contents of this post.
You want to learn even more possibilities to deal with NAs in R? Then definitely check out the following video of my YouTuber channel.
In the video, I provide further examples for is.na. I also speak about other functions for the handling of missing data in R data frames.
Now it’s on You!
I’ve shown you the most important ways to use the is.na R function.
However, there are hundreds of different possibilities to apply is.na in a useful way.
Do you know any other helpful applications? Or do you have a question about the usage of is.na in a specific scenario?
Don’t hesitate to let me know in the comments!
Appendix
The header graphic of this page illustrates NA values in our data. The graphic can be produced with the following R code:
N <- 2000 # Sample size x <- runif(N) # Uniformly distributed variables y <- runif(N) x_NA <- runif(50) # Random NAs y_NA <- runif(50) par(bg = "#1b98e0") # Set background color par(mar = c(0, 0, 0, 0)) # Remove space around plot pch_numb <- as.character( # Specify plotted numbers round(runif(N, 0, 9))) plot(x, y, # Plot cex = 2, pch = pch_numb, col = "#353436") text(x_NA, y_NA, cex = 2, # Add NA values to plot "NA", col = "red") points(x[1:500], y[1:500], # Overlay NA values with numbers cex = 2, pch = pch_numb, col = "#353436")







18 Comments. Leave new
I’m not sure why ‘The is.na Function in R (Basics)’ syntax doesn’t lead to the example showing “NAs” in the first 6 lines of the data frame. Thank you.
Hi Edward,
Thanks for the comment. Indeed, I messed something up when implementing the code on the website. Sorry for that, I just fixed it.
Regards,
Joachim
Dear Joachim,
How do you replace missing date values with a specific date in a data set?
I tried: sentdate head(sentdate)
[1] NA NA NA “0007-06-20” NA
[6] NA
However, the NA values were not replaced. Do you have some advice for replacing missing dates with specific dates?
Thank you so much.
Hey Sasha,
You can replace NA values in a data frame as shown below:
Have a look here, for a similar topic: https://statisticsglobe.com/r-replace-na-with-0/
Regards
Joachim
Hi there,
I have a countdata table in R where the first row is now NA as all of the columns were “character” class but I needed all of the rows except the first row to be “integer” class. I would like to convert the class of the first row back to a “factor” class. But the other rows must remain as “integer” class. Could you please give me some advice on how to do this?
Table would look as follows:
(Blank cell) Variable 1 Variable 2 Variable 3
GeneID NA NA NA # I need the GeneID names to appear there again
Gene_name 9 10 11
Gene_name 5 8 15
Hey Justin,
In R, all values of a column usually must have the same class, so I’m not sure if this is possible.
You may convert your data to a different structure such as long format, but this strongly depends on your data and on what you want to do with the data.
I hope that helps!
Joachim
Hello Joachim,
I started learning R about four weeks ago, I return to your site frequently. Thank you for the guidance.
In my data dataset1, I had a column1 with only ‘Yes’ or NA. I did this – data[is.na(data)] <- "No", which replaced the 'NA' with 'No'. However, those with 'No' do not show up when I do the following:
ggplot(data=dataset1) + geom_bar(mapping = aes(x=Column1))
I am getting just one bar for values with 'Yes', that is all. Am I doing anything wrong?
Thank you for your time.
Andy
Hey Andy,
Thank you very much for the great feedback! Glad you find my website helpful!
Regarding your question, I replicated your code, and the following works fine for me:
Could you check if this code works for you as well?
Regards
Joachim
Hello Joachim,
Turns out, I was doing everything right. Except, my column title had parenthesis in it. While I was using ” to envelope it, R expected“. Once I changed the type of inverted commas, I started getting the desired output. C’est la vie!
Thank you for putting your time into this, nevertheless. It is because of generous folks like you that novices like me have hope. May your tribe increase!
Thanks again for the very kind words Andy! Glad to hear that you found a solution! 🙂
Dear Joachim,
Thank you so much for your instructions. Your explanation has helped me a lot.
As a novice to Rstudio myself, I was wondering if the na.omit function allows to analyse with the valid data only.
Or should I instead use a function such as filter by valid (like in SPSS).
Best regards,
Daan
Hey Daan,
Thank you very much for the kind words, it’s great to hear that you find my tutorials helpful!
This strongly depends on your specific data situation. However, in most cases missing values should be substituted using missing data imputation techniques.
You can find more information on this topic here.
Regards,
Joachim
Hi JoaChim,
Hope you are well. Your website has been such a great help to me as a novice R user!
I’m trying to sum up variable scores across individual subjects, and want it to replace NAs with the score mean only if there is 2 or less NA values within the subject variable.
this is a sample of what my code looks like:
df %>%
df %
mutate(
TotalScore = rowSums(select(df, q1,q2,q3,q4,q5,q6)))
I’m unsure of how to continue the code for conditional NA rule. Would love your advice!
Thank you so much.
best regards,
nadhrah
Hey Nadhrah,
Thank you very much for the wonderful feedback, it’s great to hear that you find my tutorials helpful!
Regarding your question: Have you considered imputing your missing values using missing data imputation techniques? Once the missing values are replaced, you may simply use this code:
Regards,
Joachim
Hi everyone
I have a dataset with 700 participants but I have more than 700 rows for repetition the number of participant.
I need to find sum(is.na(data$ col) depend on other col years with 2003,2006,2009 and so on. To make summary for numbers of missing data and their percentage in each col if year ==2003, or if year==2006
Should I reshape it to count is.na in 700 participants or I need to do loop ?
Hey,
Thanks for the interesting question! It has inspired me to create a new tutorial on how to count missing values by groups in a data frame. You can find the tutorial here.
I hope that helps!
Joachim
Hey Joachim,
I’m trying to reclass values for a dataframe and I’m populating values in an already existing table with new values in a specific column with the ifelse function. I want to use the is.na and define what I want it to do if it finds an NA value. I want the value to stay as NA if it’s true and to report the observed value if it is false.
Here’s my code so you can understand what I am working with:
ordered_levels_ipums = c(“High school diploma or the equivalent, such as GED”, “Some college but no degree”, “Associate degree in college”, “Bachelor’s degree”,
“Master’s, professional school, or doctoral degree”)
ses_data_reduced$EDUCD_MOM_reclass<- NULL #create empty field.
ses_data_reduced$EDUCD_MOM_reclass <- ifelse(ses_data_reduced$EDUCD_MOM = 65 & ses_data_reduced$EDUCD_MOM= 81 & ses_data_reduced$EDUCD_MOM= 101 & ses_data_reduced$EDUCD_MOM= 114 & ses_data_reduced$EDUCD_MOM<= 116, "Master's, professional school, or doctoral degree",
ifelse(is.na(ses_data_reduced$EDUCD_MOM)
))))))
Hey Samuel,
I’m not exactly sure what you are trying to do. However, it seems like you are looking for something like this?
I hope this code helps to solve your problem.
Regards,
Joachim