Confirmatory Factor Analysis in R (Example)
In this tutorial, you’ll learn how to test the measurement validity of a questionnaire by performing Confirmatory Factor Analysis (CFA) in R.
The article consists of the following content:
Let’s walk through the steps of conducting CFA using R!
Packages & Sample Data
In order to use the relevant R functions, first, we need to install and load some R packages.
install.packages("psych") # Install psych package library("psych") # Load psych package install.packages("lavaan") # Install & load lavaan package library("lavaan") install.packages("semPlot") # Install & load semPlot package library("semPlot")
As seen, we need the psych, lavaan, and semPlot packages. We need the psych package mainly for importing our sample data. We will use lavaan for performing CFA, and we will visualize the result benefitting from semPlot.
We’ll utilize a dataset comprised of questions designed to measure the Big Five personality traits: Agreeableness, Conscientiousness, Extraversion, Neuroticism, and Openness. It is a built-in dataset in the psych package called bfi. Without further ado, let’s load our sample data.
data(bfi) # load dataset
In bfi, the first 25 columns represent personality self-report items (6-point response scale from 1:Very Inccurate to 6: Very Accurate), and the last three are demographic variables.
Since we are only interested in the measurement validity of the questions measuring the Big Five, we will omit the last three columns as follows.
mydata <- bfi[, 1:25] # use only 25 personality items head(mydata) # print mydata # A1 A2 A3 A4 A5 C1 C2 C3 C4 C5 E1 E2 E3 E4 E5 N1 N2 N3 N4 N5 O1 O2 O3 O4 O5 # 61617 5 4 3 4 4 2 3 3 3 3 4 4 3 4 4 3 4 2 2 3 3 1 3 4 4 # 61618 5 4 5 2 5 5 4 4 4 3 6 6 6 4 3 3 3 3 5 5 4 5 4 3 4 # 61620 2 4 5 4 4 4 5 4 5 2 5 3 4 4 5 4 5 4 2 3 4 5 5 5 5 # 61621 3 4 6 5 5 4 4 3 2 2 2 4 4 4 4 2 5 2 4 1 3 4 4 3 2 # 61622 5 3 3 4 5 4 4 5 4 5 5 5 5 4 5 2 3 4 4 3 3 4 4 3 4 # 61623 1 6 5 6 5 6 6 6 6 4 5 6 6 5 6 3 5 2 2 3 4 4 5 6 6
Items in our dataset are named in a way that corresponds with the personality traits they are intended to measure. Specifically, the first letter of each item name matches the initial of the respective personality trait. For example, items named A1, A2, A3, A4, and A5 are designed to measure Agreeableness.
If you check the documentation of the dataset, you will see that the some of the questions measuring the same trait might be reverse-worded. For instance, while E2 corresponds “Find it difficult to approach others.”, E4 is “Make friends easily.” although both measure the Extraversion trait.
This implies that lower points of E2 reflect a higher level of Extraversion, whereas higher points of E4 reflect a higher level of Extraversion. When performing CFA, responses to reverse-worded items are often recoded to ensure consistency in interpreting higher scores as reflecting higher levels of the factor being measured.
Let’s recode the reverse-worded items!
max_score = 6 # set max score var_to_be_reverted <- c("E1", "E2", "O2", "O5", "A1", "C4", "C5") # to be reverted variables mydata[var_to_be_reverted] <- lapply(mydata[var_to_be_reverted], # modify data function(x) (max_score + 1) - x)
First, we have defined the maximum score. Then we created a vector including the reverse-worded item names. Finally, we have recoded these items using the formula (max_score + 1) - x via the lapply function.
Now, we are ready to conduct CFA. Let’s do it!
Example: Confirmatory Factor Analysis
Parameter Estimation
First, we need to define our model structure. In other words, we need to specify the relationship between the latent and observed variables. For a terminological explanation, see Introduction to Factor Analysis.
# set model model <- 'Neuroticism =~ N1 + N2 + N3 + N4 + N5 Extraversion =~ E1 + E2 + E3 + E4 + E5 Openness =~ O1 + O2 + O3 + O4 + O5 Agreeableness =~ A1 + A2 + A3 + A4 + A5 Conscientiousness =~ C1 + C2 + C3 + C4 + C5 '
In lavaan syntax, =~ refers that the variables on the right hand side (observed variables) are explained by the variables on the left hand side (latent variables). In other words, the latent variable on the left is presumed to be the common underlying factor for the observed variables on the right.
You can see that the latent variables are named after what the respective questions intend to measure. For example, since the questions N1, N2, N3, N4 and N5 are designed to measure the level of Neuroticism, the latent factor is called Neuroticism.
The next step is fitting this model to our dataset to test if the defined model adequately represents the underlying relationships among the observed variables, which implies that the questions measure what they intend to measure.
Without further ado, let’s fit our model!
fit <- cfa(model, # compute CFA data = mydata, estimator ="MLR", missing = "ML")
You can see in the previous code snippet that the cfa function was utilized for the implementation. We have parsed the selected model, data, estimator and the missingness handling method. We opted for the MLR estimator (a robust ML estimator) since our data violates the normality assumption as being ordinal.
The cfa function offers some handling missingness methods. Under the assumption of MCAR or MAR, the likelihood function can be utilized as we did above. You can visit my tutorial Exploratory Factor Analysis in R how there is evidence that the missingness is at random (MCAR) in this bfi dataset.
For other missingness handling options, see lavOptions in the lavaan documentation. If you want to use multiple imputation as a handling method, you can utilize the semList function.
Let’s now print the summary of the CFA results!
summary(fit, # analysis summary fit.measures = TRUE, standardized = TRUE) # lavaan 0.6.16 ended normally after 93 iterations # # Estimator ML # Optimization method NLMINB # Number of model parameters 85 # # Number of observations 2800 # Number of missing patterns 87 # # Model Test User Model: # Standard Scaled # Test Statistic 4674.263 4047.736 # Degrees of freedom 265 265 # P-value (Chi-square) 0.000 0.000 # Scaling correction factor 1.155 # Yuan-Bentler correction (Mplus variant) # # Model Test Baseline Model: # # Test statistic 20010.482 16771.160 # Degrees of freedom 300 300 # P-value 0.000 0.000 # Scaling correction factor 1.193 # # User Model versus Baseline Model: # # Comparative Fit Index (CFI) 0.776 0.770 # Tucker-Lewis Index (TLI) 0.747 0.740 # # Robust Comparative Fit Index (CFI) 0.777 # Robust Tucker-Lewis Index (TLI) 0.748 # # Loglikelihood and Information Criteria: # # Loglikelihood user model (H0) -114278.379 -114278.379 # Scaling correction factor 1.151 # for the MLR correction # Loglikelihood unrestricted model (H1) -111941.247 -111941.247 # Scaling correction factor 1.154 # for the MLR correction # # Akaike (AIC) 228726.757 228726.757 # Bayesian (BIC) 229231.434 229231.434 # Sample-size adjusted Bayesian (SABIC) 228961.360 228961.360 # # Root Mean Square Error of Approximation: # # RMSEA 0.077 0.071 # 90 Percent confidence interval - lower 0.075 0.070 # 90 Percent confidence interval - upper 0.079 0.073 # P-value H_0: RMSEA <= 0.050 0.000 0.000 # P-value H_0: RMSEA >= 0.080 0.007 0.000 # # Robust RMSEA 0.077 # 90 Percent confidence interval - lower 0.075 # 90 Percent confidence interval - upper 0.079 # P-value H_0: Robust RMSEA <= 0.050 0.000 # P-value H_0: Robust RMSEA >= 0.080 0.016 # # Standardized Root Mean Square Residual: # # SRMR 0.072 0.072 # # Parameter Estimates: # # Standard errors Sandwich # Information bread Observed # Observed information based on Hessian # # Latent Variables: # Estimate Std.Err z-value P(>|z|) Std.lv Std.all # Neuroticism =~ # N1 1.000 1.274 0.811 # N2 0.955 0.017 56.739 0.000 1.217 0.798 # N3 0.907 0.029 31.455 0.000 1.156 0.721 # N4 0.703 0.033 21.428 0.000 0.896 0.571 # N5 0.643 0.030 21.169 0.000 0.819 0.506 # Extraversion =~ # E1 1.000 0.921 0.565 # E2 1.199 0.041 29.591 0.000 1.104 0.688 # E3 0.927 0.046 20.209 0.000 0.854 0.631 # E4 1.099 0.043 25.580 0.000 1.012 0.694 # E5 0.797 0.042 18.901 0.000 0.734 0.549 # Openness =~ # O1 1.000 0.636 0.564 # O2 0.960 0.080 12.021 0.000 0.611 0.390 # O3 1.392 0.082 16.947 0.000 0.886 0.726 # O4 0.461 0.053 8.681 0.000 0.294 0.240 # O5 0.953 0.071 13.408 0.000 0.606 0.457 # Agreeableness =~ # A1 1.000 0.470 0.334 # A2 1.583 0.106 14.871 0.000 0.744 0.635 # A3 2.042 0.144 14.165 0.000 0.960 0.738 # A4 1.522 0.122 12.465 0.000 0.716 0.484 # A5 1.828 0.144 12.727 0.000 0.860 0.683 # Conscientiousness =~ # C1 1.000 0.659 0.531 # C2 1.164 0.051 22.920 0.000 0.767 0.582 # C3 1.043 0.059 17.592 0.000 0.688 0.534 # C4 1.427 0.091 15.716 0.000 0.941 0.685 # C5 1.526 0.105 14.508 0.000 1.006 0.618 # # Covariances: # Estimate Std.Err z-value P(>|z|) Std.lv Std.all # Neuroticism ~~ # Extraversion -0.274 0.031 -8.791 0.000 -0.234 -0.234 # Openness -0.096 0.024 -4.042 0.000 -0.118 -0.118 # Agreeableness -0.132 0.019 -7.004 0.000 -0.220 -0.220 # Conscientisnss -0.243 0.023 -10.339 0.000 -0.289 -0.289 # Extraversion ~~ # Openness 0.264 0.024 11.116 0.000 0.451 0.451 # Agreeableness 0.296 0.023 12.811 0.000 0.684 0.684 # Conscientisnss 0.213 0.019 10.938 0.000 0.351 0.351 # Openness ~~ # Agreeableness 0.094 0.011 8.514 0.000 0.315 0.315 # Conscientisnss 0.126 0.017 7.500 0.000 0.300 0.300 # Agreeableness ~~ # Conscientisnss 0.106 0.012 9.186 0.000 0.341 0.341 # # Intercepts: # Estimate Std.Err z-value P(>|z|) Std.lv Std.all # .N1 2.932 0.030 98.568 0.000 2.932 1.867 # .N2 3.508 0.029 121.472 0.000 3.508 2.300 # .N3 3.217 0.030 106.141 0.000 3.217 2.008 # .N4 3.185 0.030 106.903 0.000 3.185 2.030 # .N5 2.969 0.031 96.676 0.000 2.969 1.834 # .E1 4.025 0.031 130.223 0.000 4.025 2.469 # .E2 3.857 0.030 126.938 0.000 3.857 2.403 # .E3 4.001 0.026 156.144 0.000 4.001 2.960 # .E4 4.421 0.028 160.305 0.000 4.421 3.034 # .E5 4.417 0.025 174.631 0.000 4.417 3.309 # .O1 4.816 0.021 224.959 0.000 4.816 4.265 # .O2 4.287 0.030 144.955 0.000 4.287 2.739 # .O3 4.436 0.023 191.469 0.000 4.436 3.634 # .O4 4.892 0.023 211.524 0.000 4.892 4.007 # .O5 4.510 0.025 179.166 0.000 4.510 3.396 # .A1 4.587 0.027 172.019 0.000 4.587 3.259 # .A2 4.805 0.022 216.449 0.000 4.805 4.101 # .A3 4.606 0.025 186.932 0.000 4.606 3.541 # .A4 4.700 0.028 167.727 0.000 4.700 3.178 # .A5 4.561 0.024 191.581 0.000 4.561 3.627 # .C1 4.503 0.024 191.500 0.000 4.503 3.629 # .C2 4.371 0.025 175.010 0.000 4.371 3.317 # .C3 4.303 0.024 176.233 0.000 4.303 3.341 # .C4 4.448 0.026 170.783 0.000 4.448 3.237 # .C5 3.703 0.031 120.117 0.000 3.703 2.275 # Neuroticism 0.000 0.000 0.000 # Extraversion 0.000 0.000 0.000 # Openness 0.000 0.000 0.000 # Agreeableness 0.000 0.000 0.000 # Conscientisnss 0.000 0.000 0.000 # # Variances: # Estimate Std.Err z-value P(>|z|) Std.lv Std.all # .N1 0.843 0.049 17.041 0.000 0.843 0.342 # .N2 0.846 0.047 18.040 0.000 0.846 0.363 # .N3 1.231 0.051 24.156 0.000 1.231 0.480 # .N4 1.660 0.057 29.196 0.000 1.660 0.674 # .N5 1.949 0.057 34.452 0.000 1.949 0.744 # .E1 1.811 0.058 31.495 0.000 1.811 0.681 # .E2 1.358 0.057 23.800 0.000 1.358 0.527 # .E3 1.099 0.043 25.309 0.000 1.099 0.601 # .E4 1.099 0.048 23.079 0.000 1.099 0.518 # .E5 1.244 0.044 28.312 0.000 1.244 0.698 # .O1 0.870 0.036 24.496 0.000 0.870 0.682 # .O2 2.076 0.066 31.435 0.000 2.076 0.848 # .O3 0.705 0.052 13.562 0.000 0.705 0.473 # .O4 1.405 0.051 27.280 0.000 1.405 0.942 # .O5 1.396 0.056 25.100 0.000 1.396 0.792 # .A1 1.760 0.054 32.399 0.000 1.760 0.888 # .A2 0.819 0.040 20.483 0.000 0.819 0.596 # .A3 0.771 0.043 17.790 0.000 0.771 0.455 # .A4 1.675 0.057 29.432 0.000 1.675 0.766 # .A5 0.843 0.044 19.330 0.000 0.843 0.533 # .C1 1.105 0.051 21.583 0.000 1.105 0.718 # .C2 1.148 0.049 23.239 0.000 1.148 0.661 # .C3 1.186 0.042 28.147 0.000 1.186 0.715 # .C4 1.003 0.053 18.990 0.000 1.003 0.531 # .C5 1.638 0.066 24.750 0.000 1.638 0.618 # Neuroticism 1.623 0.066 24.755 0.000 1.000 1.000 # Extraversion 0.848 0.060 14.086 0.000 1.000 1.000 # Openness 0.405 0.035 11.734 0.000 1.000 1.000 # Agreeableness 0.221 0.031 7.184 0.000 1.000 1.000 # Conscientisnss 0.435 0.043 10.161 0.000 1.000 1.000
As seen, the cfa function provides an extensive output with fit measures (TLI, CFI, etc.) and parameter estimates (factor loadings, variance, etc.) including the standardized forms thanks to the arguments fit.measures=TRUE and standardized=TRUE.
Estimate Plausibility Check
Three types of estimates are provided, the raw estimates under the estimate column Estimate, the estimates when the latent factor is standardized Std.lv and the estimates when both observed and latent variables are standardized Std.all.
It is better to check first if the parameter estimates are plausible, see the following points.
- The correlations must be between -1 and 1, see the
Std.allcolumn in theCovariancesoutput. - The variances must be non-negative, see the
Variancesoutput. - The standardized factor loadings shouldn’t be too low (commonly above 0.3 or 0.4 are acceptable), see the
Std.allcolumn in theLatent Variablesoutput. Otherwise, it implies that the question does not measure the intended concept effectively and could be nearly as uninformative as a random question. - The correlations between the latent variables shouldn’t be exclusively high (commonly above 0.70 or 0.80 are undesirable), see the
Std.allcolumn underCovariances. Otherwise, it implies that the two latent factors measure very similar concepts, rendering them indistinct or not clearly distinguishable.
In light of these points, we can conclude that all estimates are plausible except for the factor loading of O4, which implies that the question does not measure the Opennes trait as effectively as others do.
The item explanation should be revisited and compared with the other item explanations aiming to measure the same trait. If the difference holds both theoretical and practical implications, the item should be removed from the model, and the evaluation should proceed with the updated model.
Model Respecification
Assuming we can justify the removal of 04, we will respecify our model. See the following script.
# respecify model model_respecified <- ' Neuroticism =~ N1 + N2 + N3 + N4 + N5 Extraversion =~ E1 + E2 + E3 + E4 + E5 Openness =~ O1 + O2 + O3 + O5 Agreeableness =~ A1 + A2 + A3 + A4 + A5 Conscientiousness =~ C1 + C2 + C3 + C4 + C5 '
The model was redefined and named as model_respecified. The next step is fitting the respecified model to our dataset.
fit_respecified <- cfa(model_respecified, # recompute CFA data = mydata, estimator = "MLR", missing = "ML")
We haven’t changed any fit settings as seen. Now, we will print the respecified model fit summary as follows.
summary(fit_respecified, # analysis summary fit.measures = TRUE, standardized = TRUE) # lavaan 0.6.16 ended normally after 92 iterations # # Estimator ML # Optimization method NLMINB # Number of model parameters 82 # # Number of observations 2800 # Number of missing patterns 83 # # Model Test User Model: # Standard Scaled # Test Statistic 4226.203 3657.387 # Degrees of freedom 242 242 # P-value (Chi-square) 0.000 0.000 # Scaling correction factor 1.156 # Yuan-Bentler correction (Mplus variant) # # Model Test Baseline Model: # # Test statistic 19453.245 16271.394 # Degrees of freedom 276 276 # P-value 0.000 0.000 # Scaling correction factor 1.196 # # User Model versus Baseline Model: # # Comparative Fit Index (CFI) 0.792 0.786 # Tucker-Lewis Index (TLI) 0.763 0.756 # # Robust Comparative Fit Index (CFI) 0.793 # Robust Tucker-Lewis Index (TLI) 0.764 # # Loglikelihood and Information Criteria: # # Loglikelihood user model (H0) -109823.452 -109823.452 # Scaling correction factor 1.143 # for the MLR correction # Loglikelihood unrestricted model (H1) -107710.350 -107710.350 # Scaling correction factor 1.152 # for the MLR correction # # Akaike (AIC) 219810.904 219810.904 # Bayesian (BIC) 220297.769 220297.769 # Sample-size adjusted Bayesian (SABIC) 220037.227 220037.227 # # Root Mean Square Error of Approximation: # # RMSEA 0.077 0.071 # 90 Percent confidence interval - lower 0.075 0.069 # 90 Percent confidence interval - upper 0.079 0.073 # P-value H_0: RMSEA <= 0.050 0.000 0.000 # P-value H_0: RMSEA >= 0.080 0.004 0.000 # # Robust RMSEA 0.077 # 90 Percent confidence interval - lower 0.075 # 90 Percent confidence interval - upper 0.079 # P-value H_0: Robust RMSEA <= 0.050 0.000 # P-value H_0: Robust RMSEA >= 0.080 0.009 # # Standardized Root Mean Square Residual: # # SRMR 0.068 0.068 # # Parameter Estimates: # # Standard errors Sandwich # Information bread Observed # Observed information based on Hessian # # Latent Variables: # Estimate Std.Err z-value P(>|z|) Std.lv Std.all # Neuroticism =~ # N1 1.000 1.274 0.812 # N2 0.955 0.017 56.739 0.000 1.217 0.798 # N3 0.907 0.029 31.509 0.000 1.155 0.721 # N4 0.702 0.033 21.462 0.000 0.895 0.571 # N5 0.643 0.030 21.174 0.000 0.819 0.506 # Extraversion =~ # E1 1.000 0.920 0.564 # E2 1.199 0.040 29.625 0.000 1.104 0.688 # E3 0.930 0.046 20.263 0.000 0.856 0.633 # E4 1.096 0.043 25.639 0.000 1.008 0.692 # E5 0.799 0.042 18.931 0.000 0.736 0.551 # Openness =~ # O1 1.000 0.631 0.559 # O2 0.979 0.082 11.887 0.000 0.618 0.395 # O3 1.389 0.083 16.713 0.000 0.877 0.718 # O5 0.939 0.072 13.066 0.000 0.593 0.446 # Agreeableness =~ # A1 1.000 0.470 0.334 # A2 1.584 0.107 14.868 0.000 0.744 0.635 # A3 2.043 0.144 14.158 0.000 0.960 0.738 # A4 1.525 0.122 12.464 0.000 0.717 0.485 # A5 1.829 0.144 12.720 0.000 0.859 0.683 # Conscientiousness =~ # C1 1.000 0.658 0.530 # C2 1.165 0.051 22.915 0.000 0.767 0.582 # C3 1.044 0.059 17.617 0.000 0.687 0.533 # C4 1.431 0.091 15.790 0.000 0.942 0.685 # C5 1.531 0.105 14.603 0.000 1.007 0.619 # # Covariances: # Estimate Std.Err z-value P(>|z|) Std.lv Std.all # Neuroticism ~~ # Extraversion -0.274 0.031 -8.766 0.000 -0.233 -0.233 # Openness -0.118 0.023 -5.032 0.000 -0.147 -0.147 # Agreeableness -0.132 0.019 -7.003 0.000 -0.220 -0.220 # Conscientisnss -0.243 0.023 -10.353 0.000 -0.289 -0.289 # Extraversion ~~ # Openness 0.281 0.023 11.948 0.000 0.483 0.483 # Agreeableness 0.296 0.023 12.801 0.000 0.684 0.684 # Conscientisnss 0.213 0.019 10.948 0.000 0.351 0.351 # Openness ~~ # Agreeableness 0.095 0.011 8.508 0.000 0.319 0.319 # Conscientisnss 0.131 0.017 7.797 0.000 0.315 0.315 # Agreeableness ~~ # Conscientisnss 0.106 0.011 9.181 0.000 0.341 0.341 # # Intercepts: # Estimate Std.Err z-value P(>|z|) Std.lv Std.all # .N1 2.932 0.030 98.568 0.000 2.932 1.867 # .N2 3.508 0.029 121.470 0.000 3.508 2.300 # .N3 3.217 0.030 106.140 0.000 3.217 2.008 # .N4 3.185 0.030 106.903 0.000 3.185 2.030 # .N5 2.969 0.031 96.676 0.000 2.969 1.834 # .E1 4.025 0.031 130.224 0.000 4.025 2.469 # .E2 3.857 0.030 126.937 0.000 3.857 2.403 # .E3 4.001 0.026 156.145 0.000 4.001 2.960 # .E4 4.421 0.028 160.305 0.000 4.421 3.034 # .E5 4.417 0.025 174.632 0.000 4.417 3.309 # .O1 4.816 0.021 224.948 0.000 4.816 4.265 # .O2 4.287 0.030 144.955 0.000 4.287 2.739 # .O3 4.436 0.023 191.494 0.000 4.436 3.634 # .O5 4.510 0.025 179.175 0.000 4.510 3.397 # .A1 4.587 0.027 172.018 0.000 4.587 3.259 # .A2 4.805 0.022 216.450 0.000 4.805 4.101 # .A3 4.606 0.025 186.927 0.000 4.606 3.541 # .A4 4.700 0.028 167.728 0.000 4.700 3.178 # .A5 4.561 0.024 191.581 0.000 4.561 3.627 # .C1 4.503 0.024 191.498 0.000 4.503 3.629 # .C2 4.371 0.025 175.012 0.000 4.371 3.317 # .C3 4.303 0.024 176.234 0.000 4.303 3.341 # .C4 4.448 0.026 170.785 0.000 4.448 3.237 # .C5 3.703 0.031 120.117 0.000 3.703 2.275 # Neuroticism 0.000 0.000 0.000 # Extraversion 0.000 0.000 0.000 # Openness 0.000 0.000 0.000 # Agreeableness 0.000 0.000 0.000 # Conscientisnss 0.000 0.000 0.000 # # Variances: # Estimate Std.Err z-value P(>|z|) Std.lv Std.all # .N1 0.842 0.049 17.061 0.000 0.842 0.341 # .N2 0.846 0.047 18.065 0.000 0.846 0.363 # .N3 1.231 0.051 24.184 0.000 1.231 0.480 # .N4 1.661 0.057 29.262 0.000 1.661 0.675 # .N5 1.948 0.057 34.423 0.000 1.948 0.744 # .E1 1.812 0.058 31.496 0.000 1.812 0.682 # .E2 1.358 0.057 23.835 0.000 1.358 0.527 # .E3 1.095 0.043 25.313 0.000 1.095 0.599 # .E4 1.107 0.048 23.289 0.000 1.107 0.521 # .E5 1.241 0.044 28.256 0.000 1.241 0.696 # .O1 0.877 0.035 24.745 0.000 0.877 0.688 # .O2 2.067 0.067 31.027 0.000 2.067 0.844 # .O3 0.722 0.052 13.837 0.000 0.722 0.484 # .O5 1.412 0.055 25.715 0.000 1.412 0.801 # .A1 1.760 0.054 32.414 0.000 1.760 0.889 # .A2 0.819 0.040 20.492 0.000 0.819 0.597 # .A3 0.771 0.043 17.793 0.000 0.771 0.455 # .A4 1.674 0.057 29.408 0.000 1.674 0.765 # .A5 0.843 0.044 19.326 0.000 0.843 0.533 # .C1 1.107 0.051 21.669 0.000 1.107 0.719 # .C2 1.149 0.049 23.309 0.000 1.149 0.661 # .C3 1.187 0.042 28.154 0.000 1.187 0.716 # .C4 1.001 0.053 19.039 0.000 1.001 0.530 # .C5 1.635 0.066 24.854 0.000 1.635 0.617 # Neuroticism 1.624 0.065 24.801 0.000 1.000 1.000 # Extraversion 0.847 0.060 14.071 0.000 1.000 1.000 # Openness 0.398 0.035 11.515 0.000 1.000 1.000 # Agreeableness 0.221 0.031 7.177 0.000 1.000 1.000 # Conscientisnss 0.433 0.043 10.178 0.000 1.000 1.000
All parameter estimates became plausible. Let’s skip to the next step!
Fit Evaluation & Model Modification
The second step is to check the global goodness of fit indices, see Confirmatory Factor Analysis for further details.
Unfortunately, our model doesn’t fit well with the data based on the indices TLI < 0.95 , CFI < 0.95, RMSEA > 0.05. Please see the values under the column Scaled, not Standard, for the robust fit indices.
In such cases, the common next step is trying to improve the fit by freeing the model constraints.
To establish model identification, enhance parsimony, and align the model with theoretical expectations, some constraints are applied by researchers in CFA. In this example, we didn’t specified any restrisctions on parameters, yet, in lavaan, by default, the first factor loadings are fixed to 1, the measurement error (see Confirmatory Factor Analysis) covariances are fixed to 0, the cross-loadings are fixed to 0.
In the next step, we will see how freeing these default constraints will affect the goodness of fit. To do this, we will use the modificationIndices function, sorting the modifications by the expected improvement in Chi-square fit, also known as modification index.
head(modificationIndices(fit_respecified, sort = TRUE)) # check modification indices # lhs op rhs mi epc sepc.lv sepc.all sepc.nox # 189 N1 ~~ N2 442.247 0.806 0.806 0.955 0.955 # 115 Extraversion =~ N4 219.011 -0.479 -0.441 -0.281 -0.281 # 234 N3 ~~ N4 145.924 0.402 0.402 0.281 0.281 # 179 Conscientiousness =~ E5 144.899 0.535 0.352 0.264 0.264 # 139 Openness =~ E4 141.515 -0.675 -0.426 -0.293 -0.293 # 138 Openness =~ E3 136.595 0.627 0.396 0.293 0.293
We have used the head function to print only the first six rows. Because we were interested in the modifications leading fit improvement in larger amounts, which would stand on top of the sorted list by mi.
The modifications based on the expected parameter value change could also be prioritized if the user focused more on the substantive importance of freeing a parameter rather than the overall fit improvement.
This approach necessitates ordering the modifications by the expected value change, epc or the expected standardized value change sepc.all, and evaluating the top modifications. Let’s see how it can be implemented.
mi <- modificationIndices(fit_respecified) # check modification indices mi_sorted_by_epc <- mi[order(mi$sepc.all, decreasing = TRUE), ] head(mi_sorted_by_epc) # lhs op rhs mi epc sepc.lv sepc.all sepc.nox # 189 N1 ~~ N2 442.247 0.806 0.806 0.955 0.955 # 159 Agreeableness =~ E4 53.158 0.714 0.336 0.230 0.230 # 138 Openness =~ E3 136.595 0.627 0.396 0.293 0.293 # 179 Conscientiousness =~ E5 144.899 0.535 0.352 0.264 0.264 # 140 Openness =~ E5 80.817 0.490 0.309 0.232 0.232 # 125 Extraversion =~ A5 118.035 0.488 0.449 0.357 0.357
It is important to note that the modificationIndices function will only consider fixed-to-zero constraints. If you have equality constraints in the model, and you wish to examine what happens if you release these equality constraints, use the lavTestScore function.
In our case, both approaches suggest, first, freeing the error covariance between N1 and N2, indicated by ~~ in lavaan syntax, which refers to covariance.
Like earlier, the next step is to visit the item explanations and see if the suggested relation holds practical and theoretical implications. If the modification is justifiable, the model should be respecified again. See the second respecified model below.
# respecify model model_respecified_2 <- ' Neuroticism =~ N1 + N2 + N3 + N4 + N5 Extraversion =~ E1 + E2 + E3 + E4 + E5 Openness =~ O1 + O2 + O3 + O5 Agreeableness =~ A1 + A2 + A3 + A4 + A5 Conscientiousness =~ C1 + C2 + C3 + C4 + C5 N1 ~~ N2 '
As you can see, we simply added N1 ~~ N2 to a new line in the model specification. The next step is fitting the model as usual.
fit_respecified_2 <- cfa(model_respecified_2, # recompute CFA data = mydata, estimator = "MLR", missing = "ML")
We have reperformed CFA in the previous step. Let’s print the results!
summary(fit_respecified_2, # analysis summary fit.measures = TRUE, standardized = TRUE) # lavaan 0.6.16 ended normally after 93 iterations # # Estimator ML # Optimization method NLMINB # Number of model parameters 83 # # Number of observations 2800 # Number of missing patterns 83 # # Model Test User Model: # Standard Scaled # Test Statistic 3853.667 3332.798 # Degrees of freedom 241 241 # P-value (Chi-square) 0.000 0.000 # Scaling correction factor 1.156 # Yuan-Bentler correction (Mplus variant) # # Model Test Baseline Model: # # Test statistic 19453.245 16271.394 # Degrees of freedom 276 276 # P-value 0.000 0.000 # Scaling correction factor 1.196 # # User Model versus Baseline Model: # # Comparative Fit Index (CFI) 0.812 0.807 # Tucker-Lewis Index (TLI) 0.784 0.779 # # Robust Comparative Fit Index (CFI) 0.813 # Robust Tucker-Lewis Index (TLI) 0.786 # # Loglikelihood and Information Criteria: # # Loglikelihood user model (H0) -109637.184 -109637.184 # Scaling correction factor 1.141 # for the MLR correction # Loglikelihood unrestricted model (H1) -107710.350 -107710.350 # Scaling correction factor 1.152 # for the MLR correction # # Akaike (AIC) 219440.367 219440.367 # Bayesian (BIC) 219933.169 219933.169 # Sample-size adjusted Bayesian (SABIC) 219669.450 219669.450 # # Root Mean Square Error of Approximation: # # RMSEA 0.073 0.068 # 90 Percent confidence interval - lower 0.071 0.066 # 90 Percent confidence interval - upper 0.075 0.070 # P-value H_0: RMSEA <= 0.050 0.000 0.000 # P-value H_0: RMSEA >= 0.080 0.000 0.000 # # Robust RMSEA 0.073 # 90 Percent confidence interval - lower 0.071 # 90 Percent confidence interval - upper 0.075 # P-value H_0: Robust RMSEA <= 0.050 0.000 # P-value H_0: Robust RMSEA >= 0.080 0.000 # # Standardized Root Mean Square Residual: # # SRMR 0.066 0.066 # # Parameter Estimates: # # Standard errors Sandwich # Information bread Observed # Observed information based on Hessian # # Latent Variables: # Estimate Std.Err z-value P(>|z|) Std.lv Std.all # Neuroticism =~ # N1 1.000 1.048 0.667 # N2 0.949 0.024 40.334 0.000 0.995 0.652 # N3 1.213 0.036 34.152 0.000 1.271 0.794 # N4 0.992 0.042 23.635 0.000 1.040 0.663 # N5 0.861 0.037 23.286 0.000 0.903 0.558 # Extraversion =~ # E1 1.000 0.921 0.565 # E2 1.208 0.041 29.754 0.000 1.113 0.693 # E3 0.923 0.045 20.324 0.000 0.850 0.628 # E4 1.095 0.042 25.794 0.000 1.008 0.692 # E5 0.797 0.042 19.096 0.000 0.734 0.550 # Openness =~ # O1 1.000 0.631 0.559 # O2 0.976 0.082 11.876 0.000 0.616 0.394 # O3 1.392 0.084 16.591 0.000 0.878 0.719 # O5 0.939 0.072 13.072 0.000 0.592 0.446 # Agreeableness =~ # A1 1.000 0.468 0.333 # A2 1.595 0.108 14.837 0.000 0.747 0.637 # A3 2.058 0.146 14.130 0.000 0.963 0.740 # A4 1.530 0.123 12.423 0.000 0.716 0.484 # A5 1.827 0.144 12.686 0.000 0.855 0.680 # Conscientiousness =~ # C1 1.000 0.654 0.527 # C2 1.163 0.051 22.817 0.000 0.761 0.577 # C3 1.048 0.060 17.586 0.000 0.685 0.532 # C4 1.447 0.092 15.712 0.000 0.946 0.689 # C5 1.550 0.107 14.521 0.000 1.013 0.623 # # Covariances: # Estimate Std.Err z-value P(>|z|) Std.lv Std.all # .N1 ~~ # .N2 0.650 0.043 15.269 0.000 0.650 0.481 # Neuroticism ~~ # Extraversion -0.281 0.028 -9.945 0.000 -0.292 -0.292 # Openness -0.101 0.020 -4.962 0.000 -0.153 -0.153 # Agreeableness -0.101 0.016 -6.509 0.000 -0.206 -0.206 # Conscientisnss -0.218 0.021 -10.532 0.000 -0.319 -0.319 # Extraversion ~~ # Openness 0.280 0.023 11.955 0.000 0.482 0.482 # Agreeableness 0.294 0.023 12.776 0.000 0.682 0.682 # Conscientisnss 0.212 0.019 10.921 0.000 0.352 0.352 # Openness ~~ # Agreeableness 0.094 0.011 8.516 0.000 0.319 0.319 # Conscientisnss 0.130 0.017 7.731 0.000 0.314 0.314 # Agreeableness ~~ # Conscientisnss 0.104 0.011 9.138 0.000 0.341 0.341 # # Intercepts: # Estimate Std.Err z-value P(>|z|) Std.lv Std.all # .N1 2.931 0.030 98.559 0.000 2.931 1.867 # .N2 3.508 0.029 121.521 0.000 3.508 2.300 # .N3 3.217 0.030 106.144 0.000 3.217 2.008 # .N4 3.185 0.030 106.935 0.000 3.185 2.030 # .N5 2.969 0.031 96.688 0.000 2.969 1.834 # .E1 4.025 0.031 130.223 0.000 4.025 2.469 # .E2 3.857 0.030 126.941 0.000 3.857 2.403 # .E3 4.001 0.026 156.143 0.000 4.001 2.960 # .E4 4.421 0.028 160.303 0.000 4.421 3.034 # .E5 4.417 0.025 174.634 0.000 4.417 3.309 # .O1 4.816 0.021 224.949 0.000 4.816 4.265 # .O2 4.287 0.030 144.955 0.000 4.287 2.739 # .O3 4.436 0.023 191.500 0.000 4.436 3.634 # .O5 4.510 0.025 179.173 0.000 4.510 3.397 # .A1 4.587 0.027 172.019 0.000 4.587 3.259 # .A2 4.805 0.022 216.445 0.000 4.805 4.101 # .A3 4.606 0.025 186.925 0.000 4.606 3.541 # .A4 4.700 0.028 167.731 0.000 4.700 3.178 # .A5 4.561 0.024 191.578 0.000 4.561 3.627 # .C1 4.503 0.024 191.493 0.000 4.503 3.629 # .C2 4.371 0.025 175.006 0.000 4.371 3.317 # .C3 4.303 0.024 176.230 0.000 4.303 3.341 # .C4 4.448 0.026 170.784 0.000 4.448 3.237 # .C5 3.703 0.031 120.120 0.000 3.703 2.275 # Neuroticism 0.000 0.000 0.000 # Extraversion 0.000 0.000 0.000 # Openness 0.000 0.000 0.000 # Agreeableness 0.000 0.000 0.000 # Conscientisnss 0.000 0.000 0.000 # # Variances: # Estimate Std.Err z-value P(>|z|) Std.lv Std.all # .N1 1.368 0.052 26.112 0.000 1.368 0.555 # .N2 1.336 0.049 27.300 0.000 1.336 0.575 # .N3 0.950 0.051 18.474 0.000 0.950 0.370 # .N4 1.381 0.057 24.287 0.000 1.381 0.561 # .N5 1.805 0.056 32.322 0.000 1.805 0.689 # .E1 1.811 0.057 31.700 0.000 1.811 0.681 # .E2 1.338 0.057 23.662 0.000 1.338 0.519 # .E3 1.106 0.043 25.568 0.000 1.106 0.605 # .E4 1.108 0.047 23.510 0.000 1.108 0.522 # .E5 1.243 0.044 28.505 0.000 1.243 0.697 # .O1 0.877 0.036 24.699 0.000 0.877 0.688 # .O2 2.070 0.067 31.045 0.000 2.070 0.845 # .O3 0.719 0.052 13.714 0.000 0.719 0.482 # .O5 1.412 0.055 25.704 0.000 1.412 0.801 # .A1 1.762 0.054 32.464 0.000 1.762 0.889 # .A2 0.816 0.040 20.483 0.000 0.816 0.594 # .A3 0.765 0.043 17.696 0.000 0.765 0.452 # .A4 1.675 0.057 29.388 0.000 1.675 0.766 # .A5 0.850 0.044 19.534 0.000 0.850 0.538 # .C1 1.112 0.051 21.768 0.000 1.112 0.722 # .C2 1.158 0.049 23.408 0.000 1.158 0.667 # .C3 1.190 0.042 28.193 0.000 1.190 0.717 # .C4 0.993 0.052 18.968 0.000 0.993 0.526 # .C5 1.623 0.066 24.683 0.000 1.623 0.612 # Neuroticism 1.098 0.061 17.933 0.000 1.000 1.000 # Extraversion 0.848 0.060 14.178 0.000 1.000 1.000 # Openness 0.398 0.035 11.484 0.000 1.000 1.000 # Agreeableness 0.219 0.031 7.154 0.000 1.000 1.000 # Conscientisnss 0.428 0.042 10.061 0.000 1.000 1.000
Based on the global fit indices, TLI, CFI, RMSEA and SRMR, the overall fit was improved by the local modification N1~~N2. You can also see that the freed correlation is at a considerable level with 0.481.
We can also check the significance of the improvement via Chi-square difference test. In order to do that, we should call the anova function as follows.
anova(fit_respecified, # compare models fit_respecified_2) # Scaled Chi-Squared Difference Test (method = “satorra.bentler.2001”) # # lavaan NOTE: # The “Chisq” column contains standard test statistics, not the # robust test that should be reported per model. A robust difference # test is a function of two standard (not robust) statistics. # # Df AIC BIC Chisq Chisq diff Df diff Pr(>Chisq) # fit_respecified_2 241 219440 219933 3853.7 # fit_respecified 242 219811 220298 4226.2 383.17 1 < 2.2e-16 *** # --- # Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
As observed, fit_respecified_2 significantly fits better than fit_respecified. The message shown in the output just underlines that the reported chi-square statistics are standard ones since the robust difference test is a function of two standard Chi-square statistics.
It is worth noting that the Chi-square test is sensitive to sample size. In large samples, even trivial differences in fit between two models can result in a statistically significant difference. Therefore, researchers often supplement the chi-square difference test with other fit indices (e.g., differences in CFI, RMSEA) to determine the practical significance.
Considering that the global fit indices are not still at desirable levels, e.g., CFI < 0.95, the researcher may iteratively utilize the modificationIndices function to enhance the model fit as frequently as required. However, this iterative process should be approached with caution and a strong reliance on theoretical underpinnings.
In the model refinement process overfitting should be avoided. Rigorous cross-validation, using different samples to validate the modified model, could serve as an effective strategy.
Internal Consistency Check
Getting a model that fits well is not the final step. A good fit shows that our model represents the data well, but it doesn’t inherently affirm that the items are measuring the same trait consistently, also known as internal consistency or reliability.
In our model, high error variances are apparent, as seen in the std.all column within the Variances output, which is scaled from 0 to 1. This suggests a substantial variability in the question items that is not being accounted for by the trait.
This unaccounted variability can foster inconsistencies, thereby undermining the repeatability and stability of the measure across distinct samples.
To investigate the inconsistencies in trait measurements, Cronbach’s alpha will be calculated per item group that measures a common trait. We will use the alpha function by parsing the subsets of items, as each is an item group measuring one personality trait.
alpha1 <- alpha(mydata[ ,16:20]) # cronbach alpha per factor alpha2 <- alpha(mydata[ ,11:15]) alpha3 <- alpha(mydata[ ,6:10]) alpha4 <- alpha(mydata[ ,1:5]) alpha5 <- alpha(mydata[ ,21:25])
The rule of thumb, scores above 0.70 refer to acceptable internal consistency. When you print the alphas, you will see that the internal consistency for the Openness trait, alpha5, is below the threshold. Let’s print it below!
alpha5 # print alpha5 # Reliability analysis # Call: alpha(x = mydata[, 21:25]) # # raw_alpha std.alpha G6(smc) average_r S/N ase mean sd median_r # 0.6 0.61 0.57 0.24 1.5 0.012 4.6 0.81 0.23 # # 95% confidence boundaries # lower alpha upper # Feldt 0.58 0.6 0.62 # Duhachek 0.58 0.6 0.62 # # Reliability if an item is dropped: # raw_alpha std.alpha G6(smc) average_r S/N alpha se var.r med.r # O1 0.53 0.53 0.48 0.22 1.1 0.014 0.0092 0.23 # O2 0.57 0.57 0.51 0.25 1.3 0.013 0.0076 0.22 # O3 0.50 0.50 0.44 0.20 1.0 0.015 0.0071 0.20 # O4 0.61 0.62 0.56 0.29 1.6 0.012 0.0044 0.29 # O5 0.51 0.53 0.47 0.22 1.1 0.015 0.0115 0.20 # # Item statistics # n raw.r std.r r.cor r.drop mean sd # O1 2778 0.62 0.65 0.52 0.39 4.8 1.1 # O2 2800 0.65 0.60 0.43 0.33 4.3 1.6 # O3 2772 0.67 0.69 0.59 0.45 4.4 1.2 # O4 2786 0.50 0.52 0.29 0.22 4.9 1.2 # O5 2780 0.67 0.66 0.52 0.42 4.5 1.3 # # Non missing response frequency for each item # 1 2 3 4 5 6 miss # O1 0.01 0.04 0.08 0.22 0.33 0.33 0.01 # O2 0.06 0.10 0.16 0.14 0.26 0.29 0.00 # O3 0.03 0.05 0.11 0.28 0.34 0.20 0.01 # O4 0.02 0.04 0.06 0.17 0.32 0.39 0.01 # O5 0.03 0.07 0.13 0.19 0.32 0.27 0.01
The previous code script returns a very detailed output. However, traditionally, the raw_alpha, which is 0.6 in this case, is evaluated.
Nevertheless, if you are interested in which specific item performs poorly, you can check the raw_alpha column in the Realibility if an item is dropped table, which shows how the internal consistency behaves when a particular item is dropped.
For instance, we can observe that removing item O4 increases the raw_alpha value to 0.61. This may suggest that O4 negatively impacts the internal consistency of the trait measurement, highlighting that such subtle variations should prompt a deeper exploration to make a final conclusion.
Final Model Visualisation
Assuming no further refinements are necessary for our model, we can visualize the results in a path diagram. To do this, we will utilize the semPaths function of the semPlot package.
semPaths(fit_respecified_2, # path diagram of final model whatLabels = "std", intercepts = FALSE, layout = "tree", style = "lisrel")
We specified the whatLabels argument to "std" to make sure that the standardized estimates are shown, we also suppressed the paths showing intercepts, which were out of our interest. Finally, we defined design-related arguments, like layout and style. In return, the following plot was obtained.
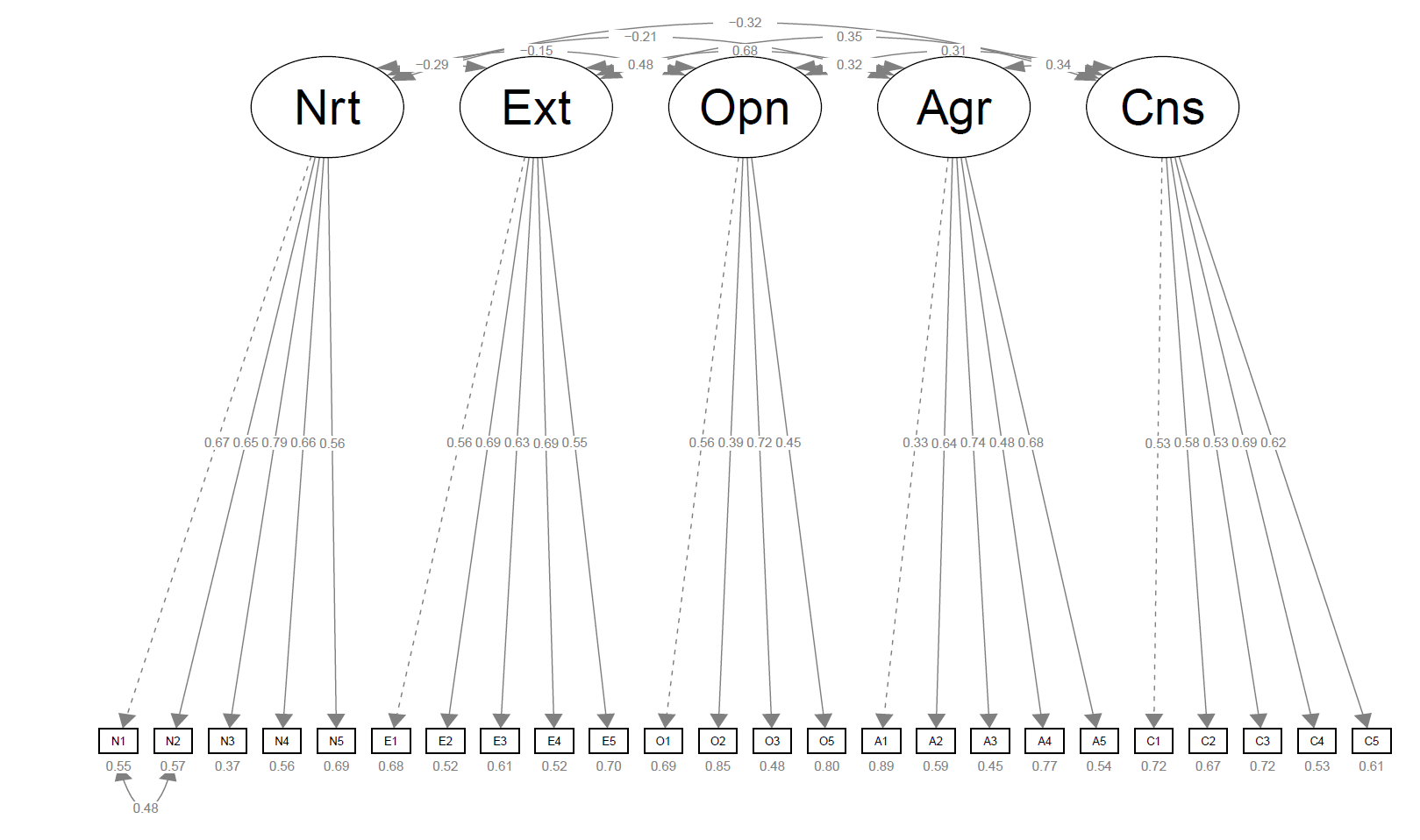
I suggest you check the function documentation for the alternative customizations. It is possible that it doesn’t provide the customization you need despite the range of options available, especially when the model is more complex.
You might explore alternative solutions for visualization in R (such as ggplot2 with ggraph) or export your model results and visualize them using general-purpose graph visualization software, like Graphviz.
Video, Further Resources & Summary
I have recently published a video on my YouTube channel, which shows the R programming codes of this tutorial. You can find the video below:
The YouTube video will be added soon.
Furthermore, you could read the related tutorials on this website:
- Intoduction to Factor Analysis
- Exploratory Factor Analysis
- Exploratory Factor Analysis in R
- Confirmatory Factor Analysis (CFA) | Meaning & Interpretation
- All R Programming Tutorials
Summary: This article has explained how to perform CFA in R in R programming. In case you have additional comments and/or questions, let me know in the comments below.
This page was created in collaboration with Cansu Kebabci. Have a look at Cansu’s author page to get more information about her professional background, a list of all her tutorials, as well as an overview on her other tasks on Statistics Globe.