Run Multiple Regression Models in for-Loop in R (Example)
In this article, I’ll show how to estimate multiple regression models in a for-loop in the R programming language.
Table of contents:
If you want to know more about these topics, keep reading…
Introducing Example Data
The following data is used as basement for this R programming tutorial:
set.seed(98274) # Creating example data y <- rnorm(1000) x1 <- rnorm(1000) + 0.2 * y x2 <- rnorm(1000) + 0.2 * x1 + 0.1 * y x3 <- rnorm(1000) - 0.1 * x1 + 0.3 * x2 - 0.3 * y data <- data.frame(y, x1, x2, x3) head(data) # Head of data # y x1 x2 x3 # 1 0.5587036 -0.3779533 -0.5320515 -0.92069263 # 2 0.8422515 -1.3835572 1.2782521 0.87967960 # 3 -0.5395343 -0.9729798 -0.1515273 -0.05973894 # 4 -0.3522260 1.2977564 -0.3512013 -0.77239810 # 5 1.5848675 -1.3152806 -2.3644414 -1.14651812 # 6 0.2207957 1.8860636 0.1967851 -0.04963894
As you can see based on the previous RStudio console output, our example data consists of four numeric columns. The first variable is our regression outcome and the three other variables are our predictors.
Example: Running Multiple Linear Regression Models in for-Loop
In this Example, I’ll show how to run three regression models within a for-loop in R. In each for-loop iteration, we are increasing the complexity of our model by adding another predictor variable to the model.
First, we have to create a list in which we will store the outputs of our for-loop iterations:
mod_summaries <- list() # Create empty list
Now, we can write a for-loop that runs multiple linear regression models as shown below:
for(i in 2:ncol(data)) { # Head of for-loop predictors_i <- colnames(data)[2:i] # Create vector of predictor names mod_summaries[[i - 1]] <- summary( # Store regression model summary in list lm(y ~ ., data[ , c("y", predictors_i)])) }
Let’s have a look at the output of our previously executed for-loop:
mod_summaries # Return summaries of all models
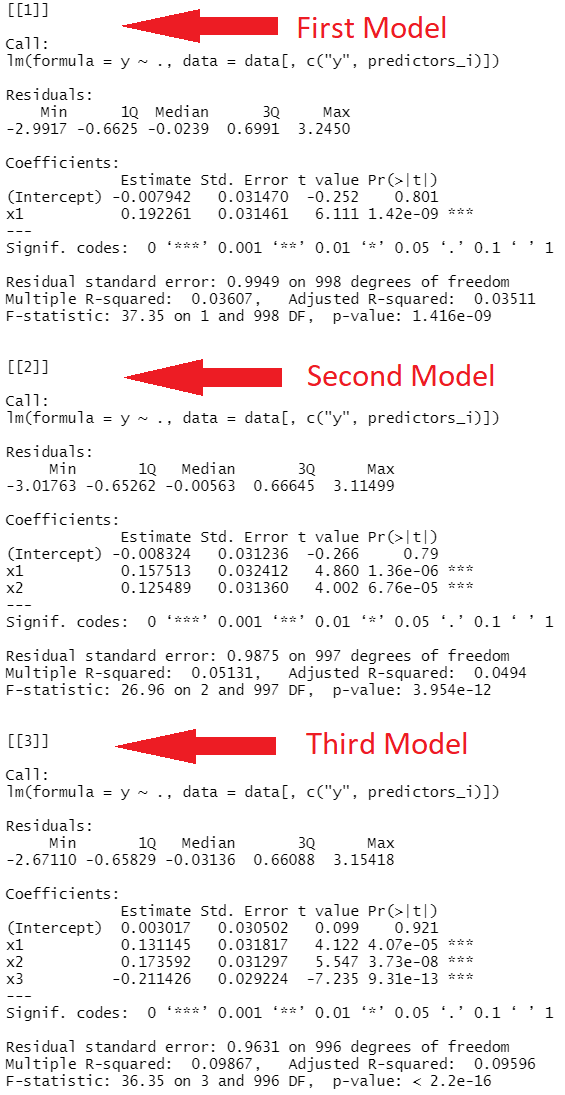
As you can see in Figure 1, we have created a list containing three different summary statistics of three different linear regressions.
Video, Further Resources & Summary
If you need further explanations on the content of this tutorial, I can recommend having a look at the following video that I have published on my YouTube channel.
In the video, I’m explaining how top loop and repeat the estimation of multiple regression models using the R programming syntax of this tutorial in RStudio.
The YouTube video will be added soon.
In addition, you may want to read the other R programming tutorials of my homepage.
- summary Function in R
- Extract Regression Coefficients of Linear Model
- for-Loop in R
- Loops in R
- The R Programming Language
Summary: At this point you should know how to write a for-loop executing several linear regressions in R programming. Please let me know in the comments below, in case you have further questions. Furthermore, please subscribe to my email newsletter to receive updates on new articles.







32 Comments. Leave new
the procedure can follow the stepwise line….
Hey Fernando,
Could you explain what you mean? I’m afraid I don’t get it.
Thanks!
Joachim
Very good
Thanks a lot Farieda, glad you like it! 🙂
Thanks for the excellent explanation. If you can provide steps on how to use PCA (principal component analysis) on series of images, that would be great!
Hey Anu,
Thanks a lot for the nice feedback, and for the topic suggestion! I’ve noted it on my to-do list.
Regards
Joachim
How can I modify this to have models for each variable by itself? For example I want the the second model to just have x2 and the third model to just have x3 so on, rather than them building onto each other. Also could I change this to do and OddsRatio with each predictor?
Hey Helen,
To change the predictor variables as you want, you only have to change
to
(i.e. remove the “2:”).
I don’t have a tutorial on Odds Ratios yet. However, I think this article explains it nicely: https://jarrettmeyer.com/2019/07/23/odds-ratio-in-r
Regards
Joachim
Hi Mr Joachim
I need your help because till now no one answered my question and I hope to find the solution with you,
How can find the range (minimum, maximum) values for Intercept, slope and R-square in r programming?
Best Regards
Hussein Ali
Hey Hussein,
Could you specify what you mean with range values?
The following tutorial shows how to extract R-squared values from a regression model: https://statisticsglobe.com/r-extract-multiple-adjusted-r-squared-from-linear-regression-model
Regards
Joachim
I mean range for Intercept, Slope and R-square, I have paper (article) they found range (minimum, maximum) for intercept, slope and r-square.
can you send to me your email to show you the paper?
I’m not an expert on this topic. However, I have recently created a Facebook discussion group where people can ask questions about R programming and statistics. Could you post your question there? This way, others can contribute/read as well: https://www.facebook.com/groups/statisticsglobe
Hello Joachim,
will this example provide me knowing which of the tested models to use for a maximised adj. R squared?
And which packages are required to run it?
Best Regards
Simon
Hey Simon,
You may combine the for-loop with the code shown in this tutorial: https://statisticsglobe.com/r-extract-multiple-adjusted-r-squared-from-linear-regression-model
This way, you could obtain a vector of R squared after running the for-loop.
You don’t have to load any additional packages for this.
Regards,
Joachim
Hi Joachim,
I’m trying to use this method to run regressions on “n” independent variables, so up to every combination available from my columns. Is there a way I can do this with this code? Thanks for your help!
Hey Alexandre,
This is definitely possible. I would find all combinations of your columns as explained here, and then I would convert each of the combinations to a formula as explained in Example 4 here.
I don’t know if there is a more efficient way, though.
Let me know in case you need further help.
Regards,
Joachim
Hi Joachim,
thank you for sharing this guide. I run several linear regression models with it. How can I extract all p-values and estimates of the list of models I created? I would like to store them in one dataframe.
Hey Simone,
Glad you find the tutorial helpful!
You may extract p-values from regression models as explained here, and you may extract regression coefficients as explained here.
Regards,
Joachim
How can we extract the predicted value from this linear models that stored in the list
Hey Foyez,
Please have a look at this tutorial for more info on this.
Regards,
Joachim
Hii Joachim,
Thanks for the nice example. How can I expand this code to extract the coeff, SD and P-value and put the in a separate list?
Hi Sabrina,
Thank you very much for the kind comment!
I just returned from holidays, so unfortunately, I couldn’t respond to your question earlier. Are you still looking for help?
Regards,
Joachim
Hii Joachim,
Yes, I am still looking for help.
Hey Sabrina,
Did you already have a look at this tutorial? It explains how to extract certain values from a regression model. You may combine this code with the code of the present tutorial.
Regards,
Joachim
Hey Joachim,
Can I modify the code and use for multiple ‘target variables’ instead of multiple ‘predictors’? If so, could you examplify the code.
Thanks
Hey Sal,
Yes, this is possible. Please have a look at the modified example code below:
I hope this helps!
Joachim
The last example is really useful for running multiple regressions in a for loop. What if I wanted to extract the regression coefficients for each i in out_names (Y1, Y2, Y3), and then transform those? For example, for each target variable (Y1, Y2, Y3) if I were interested in understanding the fraction of X1*x/Yi (such that X1*x/Yi +X2*x/Yi+X3*x/Yi = 1). Could I use a loop to extract the regression coefficient and store it so I can use it in a new function?
Hey Emily,
Thank you for the kind comment, glad you find the tutorial helpful!
Regarding your question, please have a look here. This tutorial demonstrates how to extract regression coefficients from a model. The code could be integrated in your loop.
Regards,
Joachim
Hi there, I adapted your code (thank you) for 1115 variables. Now I have a massive list with all of the summaries. Do you have recommendations on how I could easily sort through or extract only p-values or adjusted R squared values? Thanks!
Hello Nate,
Glad that we helped. To extract the adjusted Rsquares and the p-values only. You can use the following code.
For extracting the adjusted R squares:
For extracting the p-values:
Hope it helps!
Regards,
Cansu
Hello Joachim,
Could you please help me to solve this coding problem:
If I have 7 samples and I want to analyze the linear mixed model (using the LMER function) to get the variation of the antibodies logarithm according to 4 different runs.
The general code should be as follows:
Model<- lmer(log10(values)~sample + (1|Run) , data = Elisa, REML = F)
Concerning the random effect formula (1|Run): I want to get the variances for the Run variable and also the variance of the residuals for each sample (=7), how can I do this according to your explanation above.
Should I separate the sample variable into dummy variables? Or keep the sample variable as a single variable and use the loop function?
I've tried adding the Sample variable to the random effects formula like this: ((Sample|Run), but it only gives me the variances of the run without the variances of the residuals for each sample.
Thank you in advance for your help and cooperation.
And if you can write me the adjustable code, I'd be grateful.
Have a nice day.
Hello Fatme,
It has been a while since I worked with mixed models. I don’t want to guide you wrong. Besides, I am not familiar with your model and research question. I suggest you post your question on our Facebook page: https://www.facebook.com/groups/statisticsglobe with your research question. Someone with more fresh knowledge could help you there.
Best,
Cansu