Draw Histogram with Logarithmic Scale in R (3 Examples)
This article illustrates how to convert the x-axis of a graph to log scale in R.
The article is structured as follows:
It’s time to dive into the examples:
Creation of Example Data
Have a look at the following example data:
set.seed(120653) # Set random seed x <- c(rnorm(1000, 6), rnorm(1000, 13, 4)) # Create example data head(x) # Head of example data # [1] 6.717975 5.000511 5.836833 5.255884 5.894466 5.847968
The previous output of the RStudio console shows the structure of our example data – It’s a numeric vector containing randomly drawn numbers.
Example 1: Draw Histogram with Logarithmic Scale Using Base R
Example 1 shows how to create a Base R histogram with logarithmic scale.
Let’s first draw a histogram with regular values on the x-axis:
hist(x, breaks = 100) # Histogram without logarithmic axis

As shown in Figure 1, the previous R programming code created a histogram without logarithmic scale.
If we want to convert the x-axis values of our histogram to logarithmic scale, we can use the log function as shown below:
hist(log(x), breaks = 100) # Histogram with logarithmic axis

As shown in Figure 2, the previous syntax created a Base R histogram with logarithmic scale.
Example 2: Draw Histogram with Logarithmic Scale Using ggplot2 Package
In this example, I’ll explain how to draw a ggplot2 histogram with logarithmic scale.
If we want to use the functions of the ggplot2 package, we first need to install and load ggplot2:
install.packages("ggplot2") # Install & load ggplot2 package library("ggplot2")
Next, we can draw a ggplot2 histogram without log scale:
ggplot(data.frame(x), aes(x)) + # Histogram without logarithmic axis geom_histogram(bins = 100)
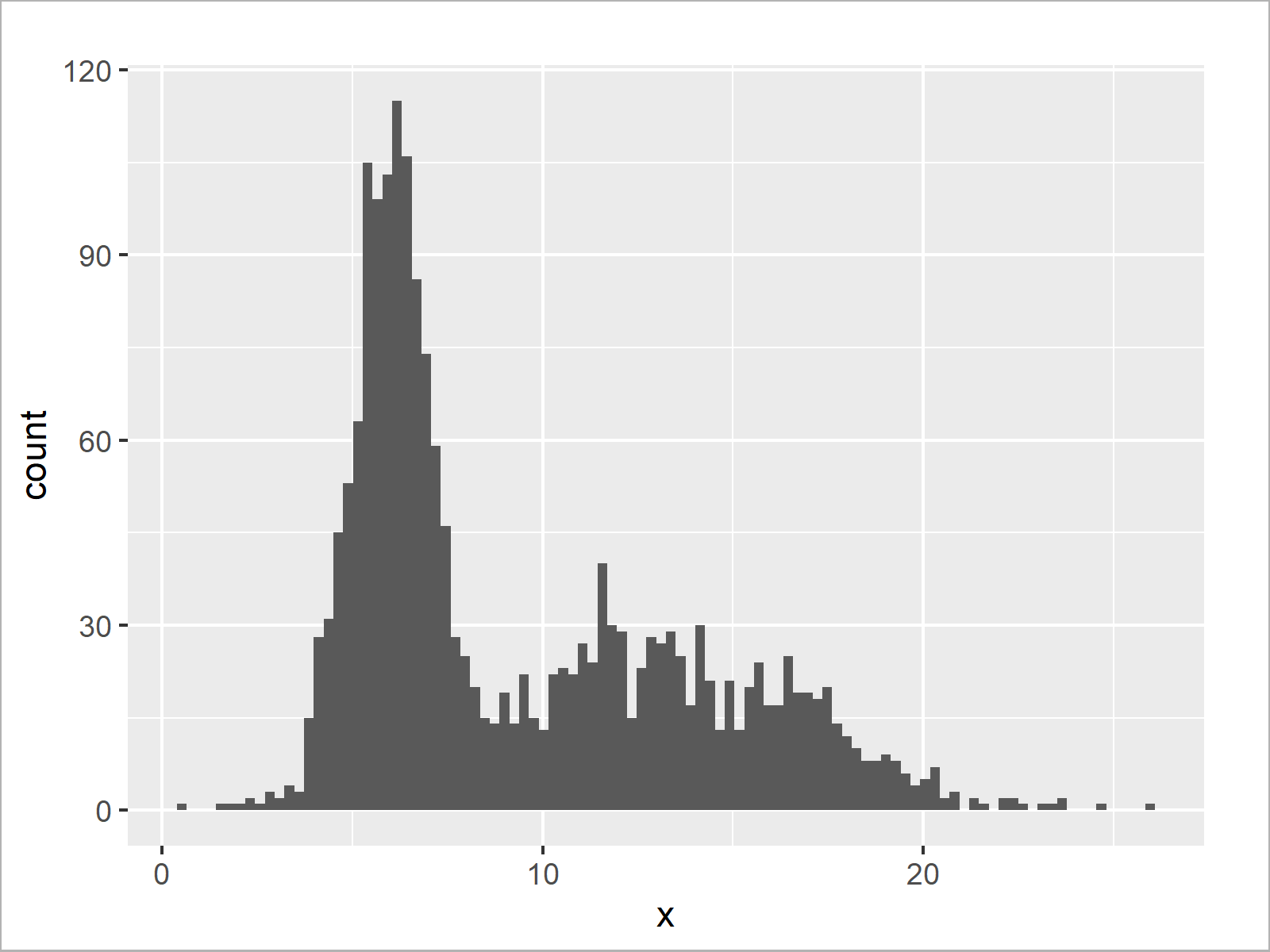
The output of the previous R code is shown in Figure 3 – A ggplot2 histogram with default x-axis values.
Let’s transform our x-axis to logarithmic scale:
ggplot(data.frame(log(x)), aes(log(x))) + # Histogram with logarithmic axis geom_histogram(bins = 100)
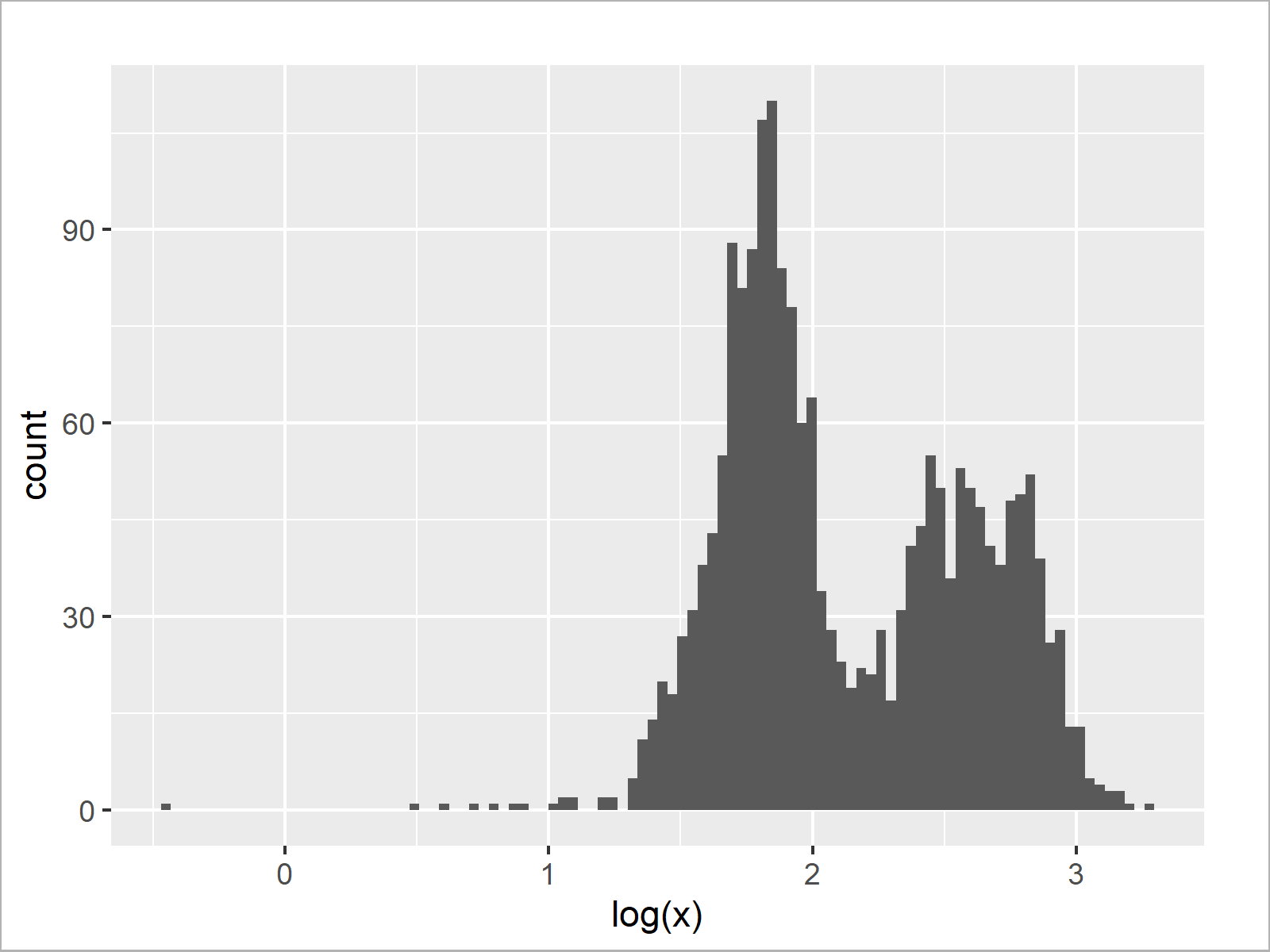
As shown in Figure 4, the previous code created a ggplot2 histogram with logarithmic scale using the log and geom_histogram functions.
Example 3: Draw Histogram with Logarithmic Scale Using scale_x_log10 Function of ggplot2 Package
In case we want to draw a ggplot2 histogram with log10 scale, we can use the scale_x_log10 function instead of the log function.
ggplot(data.frame(x), aes(x)) + # Histogram with log10 axis geom_histogram(bins = 100) + scale_x_log10()
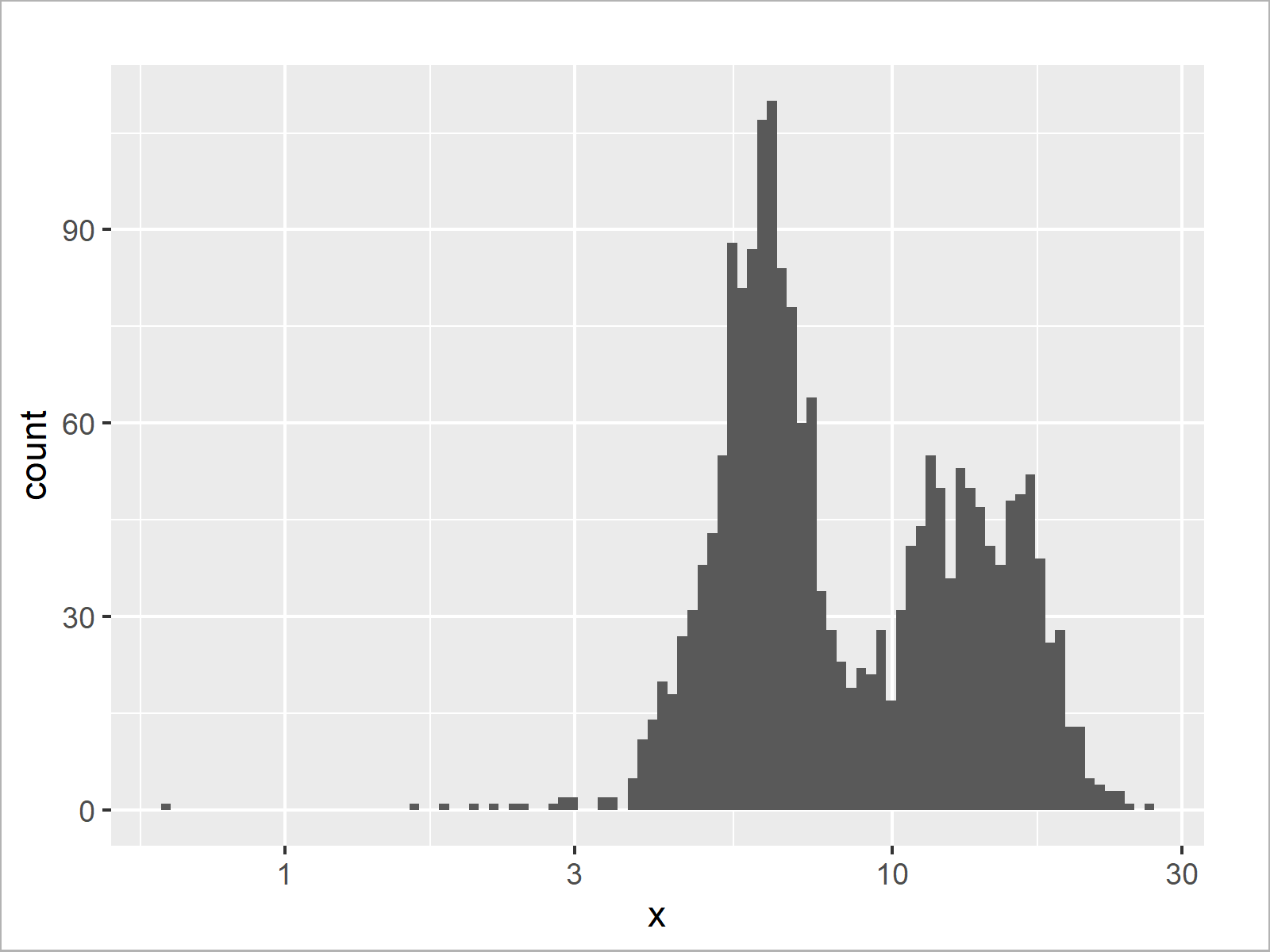
The output of the previous R programming code is shown in Figure 5 – A ggplot2 plot with log10 x-axis scale.
Video & Further Resources
Do you need more explanations on the R programming codes of this tutorial? Then you might have a look at the following video of my YouTube channel. In the video, I’m explaining the R programming code of this article:
In addition, you might want to have a look at the other articles on my website.
- Transform ggplot2 Plot Axis to log10 Scale
- Natural, Binary & Common Logarithm Using log() Function
- R Graphics Gallery
- R Programming Examples
In this post you learned how to display the x-axis of a graphic in logarithmic scale in the R programming language. In case you have any further questions, let me know in the comments section below.






2 Comments. Leave new
Hello there, could you help me with changing x-axis values for a log scaled histogram?
I have used hist(log(mydata$data, 10)) which gives me the histogram I want but now I want to change the x-axis values from log base exponents to the actual value they represent i.e., instead of 1, 2, 3, 4, 5 I’d like 10, 100, 1000, 10000, 100000.
Hello Jack,
I understand that you want to plot a histogram for the transformed data but use the actual values as labels on the x-axis. Then I suggest you run the code below.
Be aware that I also changed the x-axis label from log(x) to x.
Regards,
Cansu