Matrix Multiplication Error in R: non-conformable arguments (2 Examples)
In this article, I’ll illustrate how to deal with the error message “non-conformable arguments” in R when performing matrix multiplication.
Table of contents:
Let’s do this…
Introduction of Example Data
Have a look at the exemplifying data below:
m1 <- matrix(2) # Create first data object m1 # Print first data object
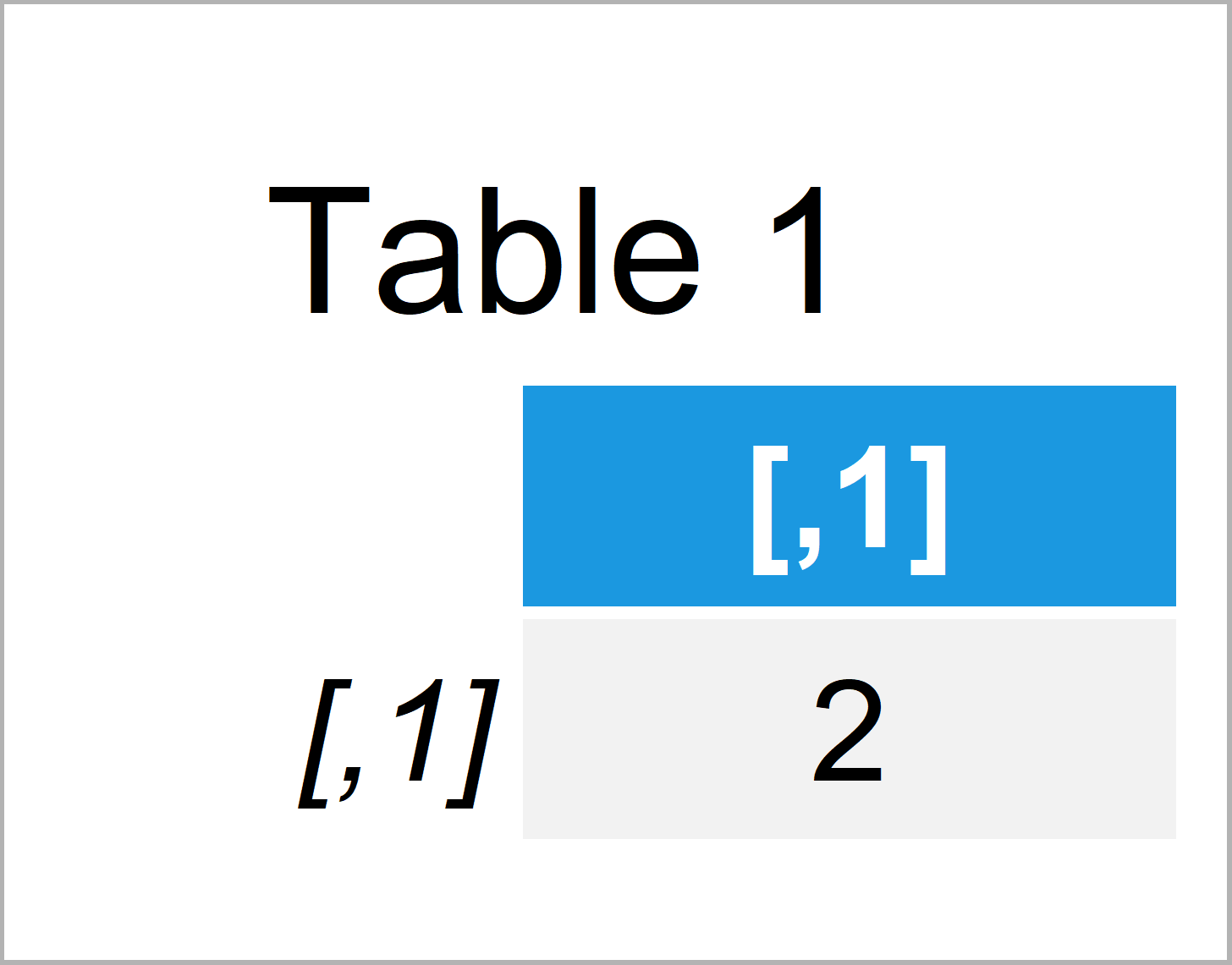
Table 1 illustrates our first data object: A matrix containing only one value.
m2 <- matrix(1:15, nrow = 5) # Create second data object m2 # Print second data object
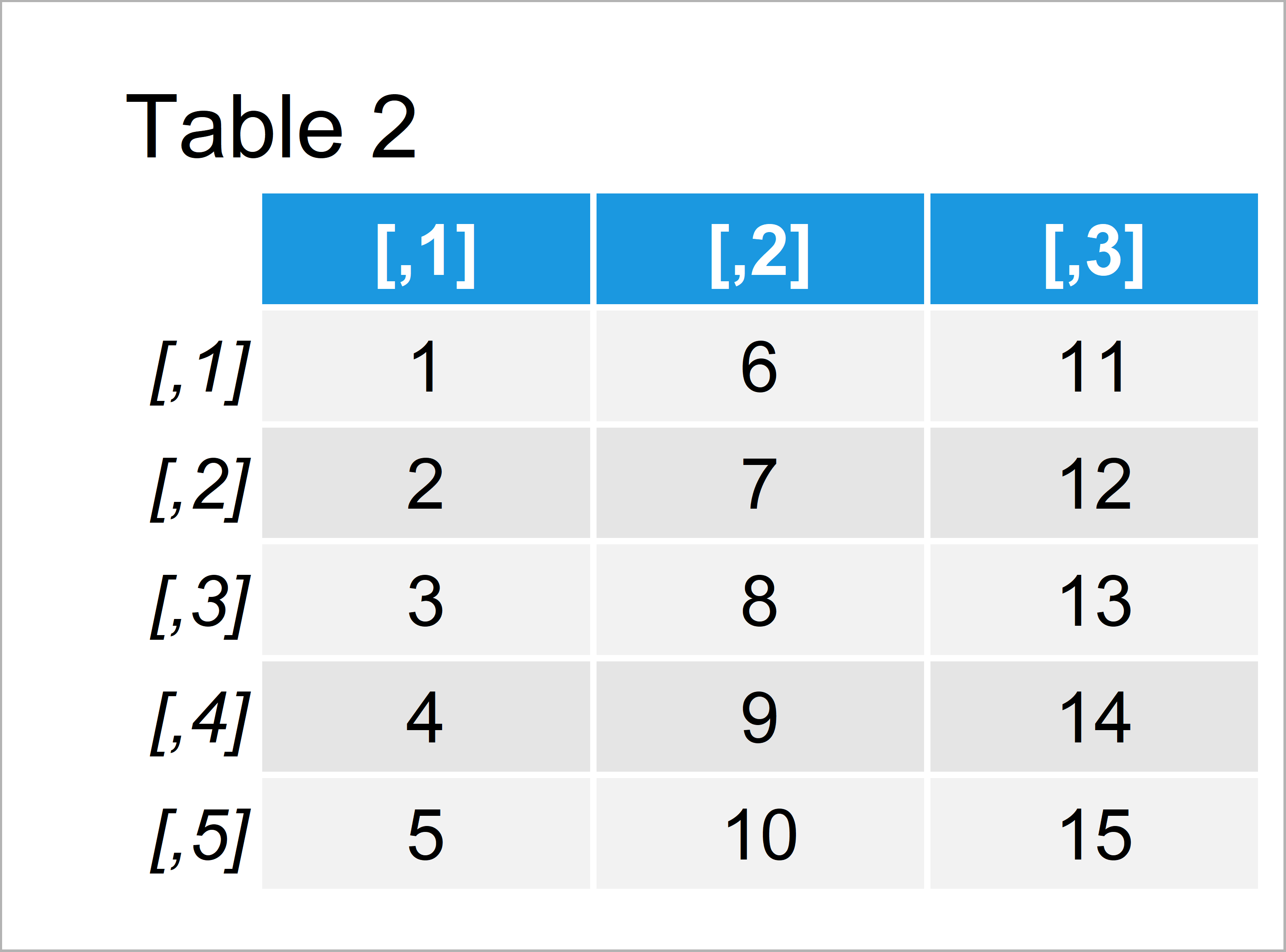
Table 2 shows the second data object: A matrix with five rows and three columns.
Example 1: Reproduce the Error Message – non-conformable arguments
In this example, I’ll explain how to reproduce the error message “non-conformable arguments” in R.
Let’s assume that we want to multiply our two data objects. Then, we might try to execute the following R code:
m1 %*% m2 # Trying to multiply data objects # Error in m1 %*% m2 : non-conformable arguments
As you can see, the error message “non-conformable arguments” has been returned to the RStudio console.
The reason for this is that our first data object m1 is a 1×1 matrix.
Let’s see how we can solve this problem…
Example 2: Fix the Error Message – non-conformable arguments
This example explains how to handle the error message “non-conformable arguments”.
To do this, we have to convert our 1×1 matrix to a vector using the as.vector function:
as.vector(m1) * m2 # Converting m1 to vector
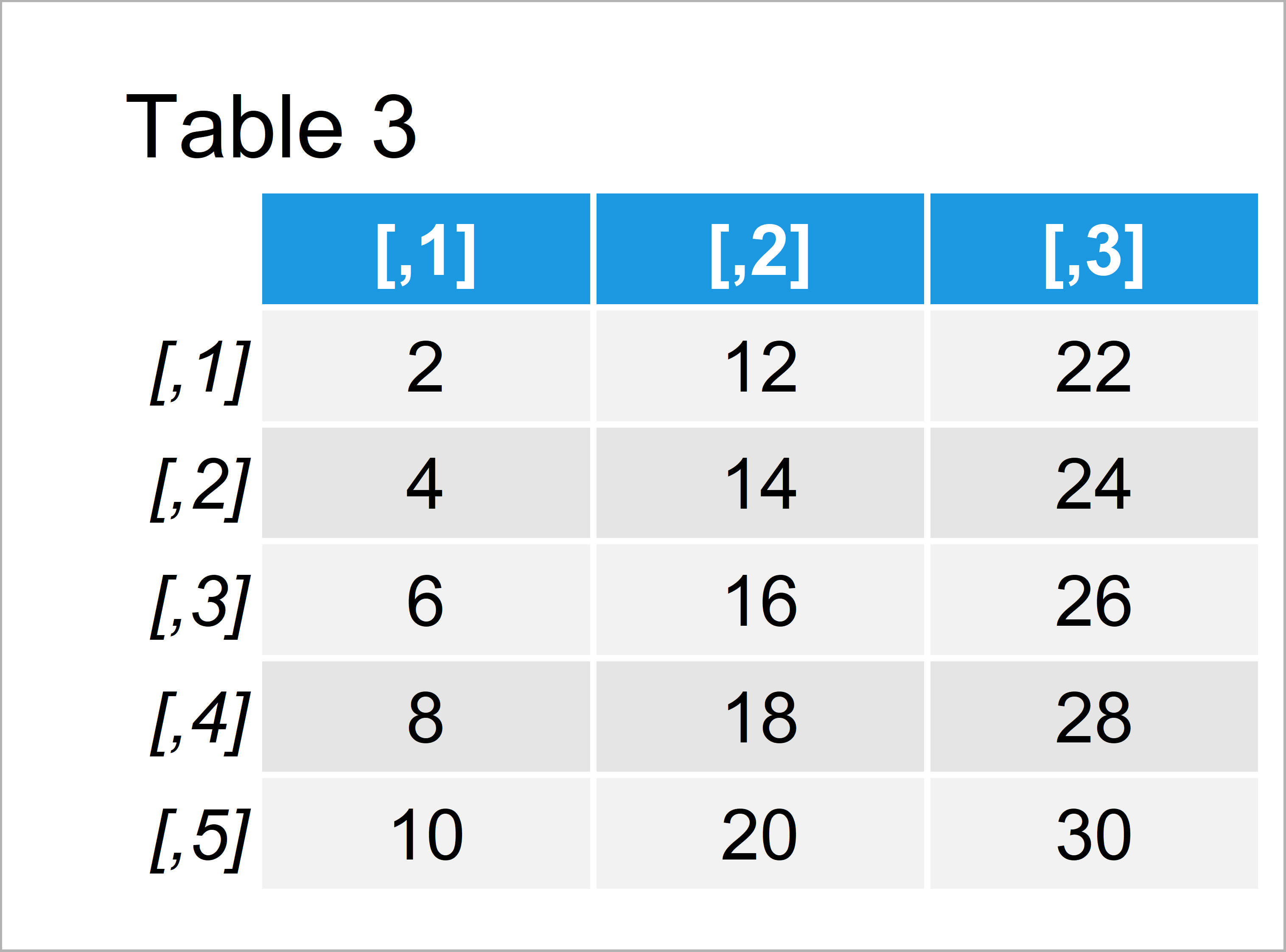
Table 3 illustrates the result of the previous R syntax.
Video & Further Resources
In case you need further information on the R programming code of this tutorial, you might want to watch the following video of my YouTube channel. In the video, I’m explaining the topics of this tutorial in R:
Furthermore, you might want to have a look at the other R tutorials on my website.
Summary: At this point you should have learned how to avoid the error “non-conformable arguments” in R programming. If you have additional questions, let me know in the comments section below.







21 Comments. Leave new
I attempted to apply lordif to examine gender differences in my measurement. My gender variable is in the first column followed by the 10 items in the measure. Total N = 1057, almost evenly split by gender. I used the following code:
genDIF <- lordif(Neur_IRT, [ , 1, criterion = 'Chisq', alpha = 0.01
I received message: unexpected '[' in genDIF <- lordif (Neur_IRT, ["
Hey Robert,
It seems like the specification of the arguments within the lordif function is incorrect. What do you want to specify with the [ ? Furthermore, it seems like a ) is missing at the end of this line of code.
Regards,
Joachim
hello,my task is Nonnegative Matrix Factorization,i also meet this problem,but in my code,it is all assumption,so i not find specific matrix to solve,so this problem need how to solve?
Hey,
Could you please share the code and data you have used?
Regards,
Joachim
Dear Joachim,
My Xp and betahatp are as follows(3*2 and 2*1), but I still cannot do Xp*betahatp, could you please help with this? Thank you in advance.
Xp:
[,1] [,2]
[1,] 2 5
[2,] 3 6
[3,] 4 7
betahatp
[,1]
[1,] 1.3333333
[2,] -0.3333333
Xp*betahatp
Error in Xp * betahatp : non-conformable arrays
Best regards,
Yina
Hey Yina,
Are you looking for the following result?
Regards,
Joachim
(my_omg) <- psych::omega(dsse_data,
nfactors = 4,
fm = "minres",
key = c(1, 1, 1, 1, -1, 1, 1, -1, -1, -1, -1, -1, 1, 1, 1, -1, -1, -1, -1, 1, -1, 1, -1, 1, 1, rep(1, 13)),
poly = TRUE
)
hey, I'm receiving the error: Error in diag(key) %*% m : non-conformable arguments
Hey Manisha,
This is difficult to evaluate without seeing the data in dsse_data. Could you illustrate how these data looks like?
Regards,
Joachim
hello, could you help me
X <- model.matrix(status~WOE_usia+WOE_penghasilan+WOE_debitrasio+WOE_keluarga+
WOE_terlambat30+WOE_terlambat60+WOE_terlambat90,
data = datascoring.WOE)
X=X[,-1]
lasso lasso <- lars(x=X,y=datascoring.WOE$status,trace = TRUE)
LASSO sequence
Computing X'X …..
Error in t(y) %*% x : non-conformable arguments
Hey Fani,
I’m not an expert on the lars package, but it seems like this Stack Overflow thread is discussing your problem. Does this help?
Regards,
Joachim
Complete the code chunk in the template to write a function my_chol that accepts a square, positive definite matrix and returns the Cholesky Decomposition in the form of a lower triangular matrix. You may not use the built-in chol() and within your function, although you can use them to validate your answers.
Your code does need to confirm that the input matrix M is square and positive definite.
Hi Ben,
Could you share the code you have tried yourself?
Regards,
Joachim
hello sir
I try to run my programing in r , but there is a problem.
x1<-rnorm(52)
x2<-rnorm(52)
x3<-rnorm(52)
x4<-rnorm(52)
x5<-rnorm(52)
y<- rnorm(52)
lm_fit<- lm(y~x1+x2+x3+x4+x5)
b<- coefficients(lm_fit)
theta<- b[c("x1","x2","x5")]
theta1<- cbind(0.007575006 , 0.008085839 ,-0.010551001)
dim(theta1)
#long run elasticities
ph2<- b[2]/(1-b[6])
cov1 <- matrix(c(-0.00021705,1.61265e-5,-0.0001109,1.61265e-5,0.0030279,-0.0021881,-0001109,
-0.0021881,0.0020943), nrow = 3 ,ncol = 3)
install.packages("msm")
library(msm)
std.err<- deltamethod(~x1/(1-x5),theta1,cov1)
Error in gdashmu %*% cov : non-conformable arguments
i do not do with this error. can you help me please?
Hey Razieh,
Please excuse the delayed response. I was on a long vacation, so unfortunately I wasn’t able to get back to you earlier. Do you still need help with your question?
Regards,
Joachim
Hi,
I hope you can help me with this. I don’t know why I’m getting a non-conformable arrays error. Here is the output
> A A
[,1] [,2] [,3]
[1,] 1 2 3
[2,] 4 5 6
> AT AT
[,1] [,2]
[1,] 1 4
[2,] 2 5
[3,] 3 6
> B B
Error: object ‘B’ not found
>
Hello Matt,
You are using the wrong operator for the matrix multiplication. See the correct one below.
Regards,
Cansu
I’m not sure why the code didn’t show. here it is as the code not the output.
A <- rbind(c(1,2,3),c(4,5,6))
A
AT <- cbind(c(1,2,3),c(4,5,6))
AT
B<- AT*A
Do you know how to calculate clustered standard errors for multinomial regression in R? I am using the ‘multinom()’ command, but it does not work well with the ‘Sandwich’ package.
Hello Nikki,
I couldn’t respond sooner, since I was on vacation. Do you still need help?
Best,
Cansu
hello. i have this function can you please help me fix this error
library(tidyverse)
library(dplyr)
library(wgeesel)
library(glmtoolbox)
library(missMethods)
generate_results <- function(seed, corstr,rho, n, each) {
set.seed(seed)
num_simulations <- 1000
results <- vector("list", num_simulations)
for (i in 1:num_simulations){
n=n
id=rep(1:n,each=each)
n_total <- n*each
rho=rho
phi=1
x=cbind(1,runif(length(id)))
beta=c(0.5,0.5)
x_mis=cbind(0,rnorm(length(id)))
para=c(0)
com.data <- data_sim(id=rep(1:n,each=each),rho=rho,phi=1,x=cbind(1,runif(length(id))),
beta=c(0.5,0.5),x_mis=cbind(0,rnorm(length(id))),para,corstr = corstr,
family = "binary",lag_level = 1)
mcar_0.05 <- delete_MCAR(com.data$data, p=0.05, cols_mis = c(6))
case1 <- select(mcar_0.05, id, response, '2')
id <- com.data$id
response <- factor(com.data$response)
'2' <- as.numeric(case1$'2')
model <- response ~ case1$'2'
if(corstr == "independence") {
Ri <- glmgee(model, id = id, data = case1, family = binomial, corstr = "independence")
} else if(corstr == "exchangeable") {
Ri <- glmgee(model, id = id, data = case1, family = binomial, corstr = "exchangeable")
} else if(corstr == "ar1") {
Ri <- glmgee(model, id = id, data = case1, family = binomial, corstr = "AR-M", Mv=1)
}
summary(Ri)
## to extract estimates
beta_o <- coef(Ri)[1] ##estimate of intercept
beta_1 <- coef(Ri)[2] ## estimte of "2"
standard_errors <- sqrt(diag(vcov(Ri)))
beta_o_rse <- standard_errors[1] ## rse of intercept
beta_1_rse <- standard_errors[2] ## rse of "2"
## extracting p-value
mull <- 0
z_stat_betao <- (beta_o – null)/beta_o_rse
p_value_betao <- 2*(1-pnorm(abs(z_stat_betao))) ## p-value for intercept
z_stat_beta1 <- (beta_1 – null)/beta_1_rse
p_value_beta1 <- 2*(1-pnorm(abs(z_stat_beta1)))
fit11 <- glmgee(model, id = id, data = case1, family = binomial) ## assumes independence
fit12 <- update(fit11, corstr="Exchangeable")
fit13 <- update(fit11, corstr="AR-M", MV=1)
cic <- CIC(fit11, fit12, fit13)
min_cic <- which.min(cic$CIC)
corr_cic <- cic$Correlation[min_cic]
qic <- QIC(fit11, fit12, fit13)
min_qic <- which.min(qic$QIC)
corr_qic <- qic$Correlation[min_qic]
rjc <- RJC(fit11, fit12, fit13)
min_rjc <- which.min(rjc$RJC)
corr_rjc <- rjc$Correlation[min_rjc]
pac <- PAC(fit11, fit12, fit13)
min_pac <- which.min(pac$PAC)
corr_pac <- pac$Correlation[min_pac]
gpc <- AGPC(fit11, fit12, fit13, k = 0)
max_gpc <- which.max(gpc$AGPC)
corr_gpc <- gpc$Correlation[max_gpc]
sgpc <- SGPC(fit11, fit12, fit13)
max_sgpc <- which.max(sgpc$SGPC)
corr_sgpc <- sgpc$Correlation[max_sgpc]
results[[i]] <- list(corr_cic = corr_cic, corr_qic = corr_qic, corr_rjc = corr_rjc, corr_pac = corr_pac,
corr_gpc = corr_gpc, corr_sgpc = corr_sgpc, beta_o = beta_o,
beta_1 = beta_1, beta_o_rse = beta_o_rse, beta_1_rse = beta_1_rse,
p_value_betao = p_value_betao, p_value_beta1 = p_value_beta1)
}
return(results)
}
## A
case2_results <- generate_results(48, "exchangeable", 0.3, 30, 3)
It was able to replicate and produce some results but along the way, this error has occured
Error in crossprod(Vi2, Xiw) : non-conformable argument
and the traceback option has displayed this
4.
crossprod(Vi2, Xiw)
3.
score(datas[[i]], beta = beta_old, out = FALSE)
2.
glmgee(model, id = id, data = case1, family = binomial, corstr = "independence")
1.
generate_results(50, "independence", 0.3, 30, 3)
i have to do thousands of simulation for different data and checking this one by one will be impractical. Thank you
Hello,
The error message you provided, Error in crossprod(Vi2, Xiw) : non-conformable argument, suggests that the two matrices or vectors Vi2 and Xiw cannot be multiplied together due to incompatible dimensions. The operation crossprod(Vi2, Xiw) is effectively equivalent to the matrix product t(Vi2) %*% Xiw, where t() is the transpose function.
Given the traceback, the error is originating from the function score which is being called by glmgee and then your generate_results function.
In order to solve the issue, for a single iteration, debug the score function and inspect the dimensions of both Vi2 and Xiw just before the crossprod function is called. Then you can see how and why the dimensions are incompatible.
Best,
Cansu