Write & Read Multiple CSV Files Using for-Loop in R (2 Examples)
In this R tutorial you’ll learn how to export and import multiple CSV files using a for-loop.
Table of contents:
Let’s take a look at some R codes in action:
Creation of Example Data
First, we’ll have to construct some exemplifying data frames in R:
data1 <- data.frame(x1 = 1:5, # First data frame x2 = letters[1:5]) data2 <- data.frame(y1 = 1:5, # Second data frame y2 = letters[1:5]) data3 <- data.frame(z1 = 1:5, # Third data frame z2 = letters[1:5])
We also have to create a directory folder on our computer were we can store our data as CSV files. For this, we can use the dir.create function as shown below:
dir.create("C:/Users/Joach/Desktop/My Folder") # Create folder
Note that you have to replace the previously used directory path by your own path. After running the previous R code you should see a new folder on your desktop.
Example 1: Writing Multiple CSV Files to Folder Using for-Loop
In this Example, I’ll show how to export multiple data frames from R to a directory using a for-loop. First, we have to specify the names of all data frames we want to export:
data_names <- c("data1", "data2", "data3") # Create vector of names data_names # Print names # "data1" "data2" "data3"
Now, we can run a for-loop that writes all our data frames to a folder using the write.csv2 function as shown below. Within the for-loop, we are specifying the names of our data frames wrapped by the get function and our directory path:
for(i in 1:length(data_names)) { # Head of for-loop write.csv2(get(data_names[i]), # Write CSV files to folder paste0("C:/Users/Joach/Desktop/My Folder/", data_names[i], ".csv"), row.names = FALSE) }
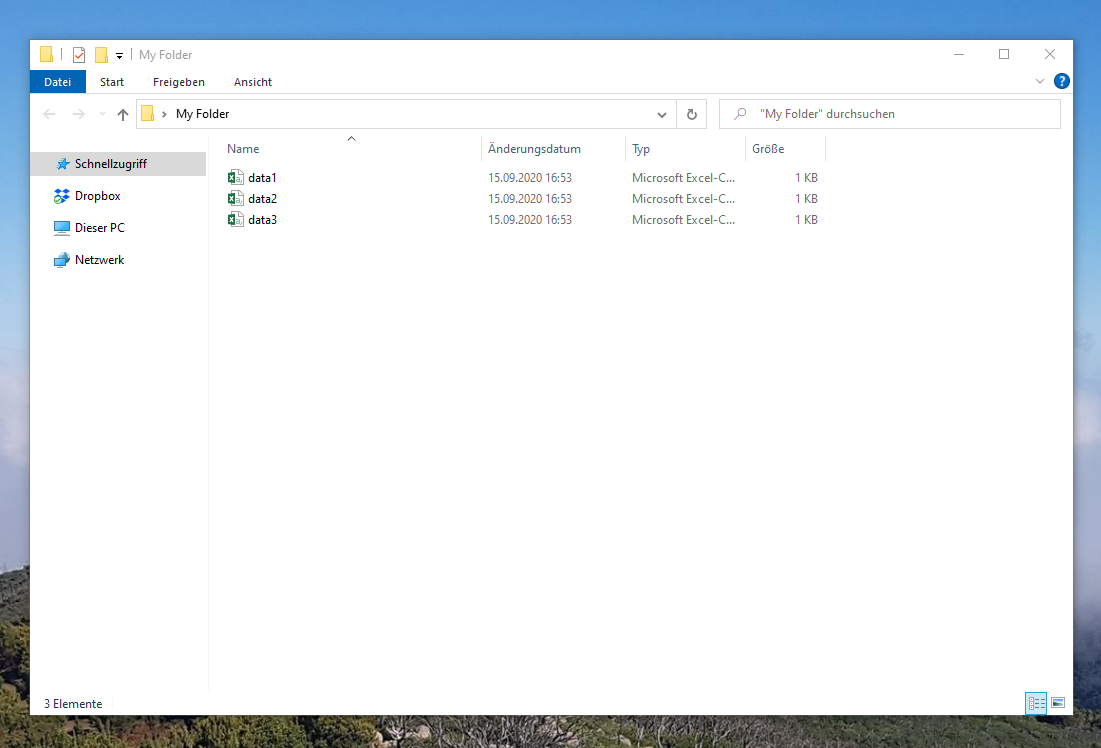
Figure 1 shows how our folder should look like after running the previous R codes. In the folder, you can see three CSV files.
Example 2: Reading Multiple CSV Files from Folder Using for-Loop
Example 2 illustrates how to import multiple CSV files using a for-loop in R. First, we have to use the list.files function to extract all file names in our folder:
data_files <- list.files("C:/Users/Joach/Desktop/My Folder") # Identify file names data_files # Print file names # "data1.csv" "data2.csv" "data3.csv"
Now, we can write a for-loop containing the assign, paste0, and read.csv2 functions to read and save all files in our directory:
for(i in 1:length(data_files)) { # Head of for-loop assign(paste0("data", i), # Read and store data frames read.csv2(paste0("C:/Users/Joach/Desktop/My Folder/", data_files[i]))) }
Our data frames are now stored in the data objects data1, data2, and data3:
data1 # Print first data frame # x1 x2 # 1 1 a # 2 2 b # 3 3 c # 4 4 d # 5 5 e data2 # Print second data frame # y1 y2 # 1 1 a # 2 2 b # 3 3 c # 4 4 d # 5 5 e data3 # Print third data frame # z1 z2 # 1 1 a # 2 2 b # 3 3 c # 4 4 d # 5 5 e
Video, Further Resources & Summary
Have a look at the following video of my YouTube channel. I show the R programming syntax of this tutorial in the video.
The YouTube video will be added soon.
Furthermore, you might want to read the other tutorials on this website. You can find some tutorials about for-loops below.
- for-Loop in R
- Loops in R
- Check in R if a Directory Exists and Create if It doesn’t
- Import & Merge Multiple csv Files
- List All Files with Specific Extension
- The R Programming Language
To summarize: This article illustrated how to read and write CSVs in loops in the R programming language. If you have further questions, don’t hesitate to let me know in the comments.







24 Comments. Leave new
Is there a way to skip lines/rows when using the example 2 method. The loop worked for me but I need to skip 13 lines so my variables are incorporated.
Hey Mason,
You mean you want to remove certain rows from your data sets? In this case, I would import the entire data sets and remove those rows afterwards within R.
Regards
Joachim
Hi Mason,
I am assuming you want to skip the first x lines.
If that is the case then include the skip = 13 in the read.csv syntax.
Regards,
Conrad
Hey Conrad,
Thanks for your help, that’s very kind!
Regards,
Joachim
how can we put separator for csv files in this files. I have many csv files with columns separated by comma
Hey Shah,
Apologies for the delayed response, I just got back from vacation. Do you still need help with this question?
Regards,
Joachim
Please can you tell me how to loop through column names and write to csv each column name. Thanks
Hey Sangay,
You may simply use the colnames function to extract all the column names from your data (i.e. colnames(data)).
Does this solve your problem?
Regards,
Joachim
thanks a lot
can we do it the same using dplyr? like map or apply?
thanks
Hey Stefano,
I’m uncertain if this is precisely what you are looking for, but this tutorial explains how to import and merge CSV files using lapply and pipes.
Regards,
Joachim
Good afternoon Joachim and Statistics Globe,
I am iterating through CSV files in a folder like the above example 2, however instead of the new name “data ith” I am trying to figure out how to dynamically assign the initial file name that appears in my list. Is there an additional step within the assign function that can do this?
Appreciate your work!
Hey Stacy,
Thanks a lot for the kind comment, glad you like our tutorials!
Regarding your question, you may replace paste0(“data”, i) by data_files[i] as shown in the following code.
Regards,
Joachim
Hi Joachim, your tutorial helped me a lot, too. I have the same question as shah initially: The data files (data1, data2, etc in your example) are not separated into columns yet. How do I add “,” as a separator, so the data files will have multiple columns?
Hi Hannah,
Thanks a lot for the kind comment, glad the tutorial is useful to you!
Could you please try to use read.csv instead of read.csv2? This might already resolve your problem.
Regards,
Joachim
Dear Joachim
i used your instruction to write multiple csv, but i got the error “object filename not found”
This is my code. ee is the names in the list files.
ee=c(files)
for(i in 1:length(ee)) { # Head of for-loop
write.csv2(get(ee[i]), # Write CSV files to folder
paste0(“D:\\”,
ee[i],
“.csv”),
row.names = FALSE)
}
}
any help would be appreciated
Hello Elham,
First of all, sorry for the late response. This error should indicate that your file path is not completely correct. Is it possible for you to provide the content of ee[i]?
Regards,
Cansu
Hi Cansu
Thank you very much for your response!
I solved the problem with
removing get and directly used the following command:
Write.csv(file=paste0(ee[i]),”.csv”)
I appreciate your reply.
Kind regards
Hey Elham,
You are welcome. Glad that you could solve it!
Regards,
Cansu
Hi,
Thanks for the easy explanation of the for loop.
I am trying to use the above example to read multiple .txt files-> transpose the table -> write it as individual .csv
data_files <- list.files("PATH")
data_files
for(i in 1:length(data_files)) { # Head of for-loop
read.table(paste0(data_files[i]))
transpose(get(data_files[i]))
write.csv2(get(data_files[i]),
paste0("PATH",
data_files[i],
".csv"))
}
But I am getting this error "Error in get(data_files[i]) : object 'B001.feature_table.txt' not found"
Could you please suggest some changes to fix this.
Thanks a lot.
Hello Kunal,
I was on vacation. That’s why I couldn’t get back to you sooner. Do you still need help? The error you’re seeing stems from the use of get(data_files[i]). The get function tries to fetch an object from the environment with the name given by its argument. However, in your case, data_files[i] returns a filename, and there’s no object with that filename in the environment. Please make sure that the file exists in the correct location.
Best,
Cansu
is it possible to import multiple csv files to respective tables in a database.I have exported the respective tables data from oracle.Now i want to import into Posgress
Hello Ajit,
I am not so familiar with this topic but these links seem helpful: Import Data from CSV into Postgres and Import CSV File Into PostgreSQL Table. If you still have trouble to solve it, then I suggest you to post your question on our Facebook discussion group.
Best,
Cansu
Hello,
I have copied the code exactly (apart from changing the file path for the newly created folder), but nothing happens when I run it. I don’t get any errors, the folder is created but it remains empty.
Any ideas what I might be doing wrong?
With thanks,
Ben
Hello Benjamin,
Could you please share your code?
Best,
Cansu