Fay-Herriot Small Area Estimator (Example in R) | Area-Level Model
In this post, we focus on the most famous area-level Small Area Estimation (SAE) model, the Fay-Herriot (FH) model. After introducing the theory behind the model, we show how to use this theory to create example data and calculate the model in the R programming language.
We cover the following blocks:
Let’s start right off with the Fay-Herriot model!
Theory of the Fay-Herriot Model
If small area estimation is completely new to you, we recommend that you first take a look at our basic article about small area estimation. In a nutshell: A small area problem is a situation where survey direct estimates are associated with high variances due to small sample sizes.
In those situations, we often prefer the use of model-based small area techniques to get more precise estimates. As these situations occur especially for the estimation in small domains, the procedure is called small area estimation.
Model-based SAE techniques can be defined at the unit- or area-level, depending on the availability of the survey and auxiliary data. When we only have access to aggregated survey information, we can only calculate area-level models like the Fay-Herriot (FH) model. The FH model is named after the article by Fay and Herriot (1979).
We have a population \(U\). For example, we consider all persons in the U.S. and are interested in the number of persons below a certain poverty threshold in the U.S. counties. Formally, we consider a total of \(D\) counties as our domains of interest. The county-specific values of the poverty parameter are denoted by \(\mu_d\), \(d=1,\ldots,D\), our parameters of interest.
To get estimates of the \({{\mu}}_d\), we conduct a survey. With the survey sample, we calculate survey direct estimates for these parameters and denote them by \(\hat{{\mu}}_d^{Dir}\). As we observe only the sample information in the \(D\) domains, the direct estimators are associated with sampling errors \({e}_d\).
Formally, this sampling model is given by
\begin{equation}
\hat{{\mu}}_d^{Dir} = {{\mu}}_d + {{e}}_d,\quad d=1,\ldots,D,
\end{equation}
where \({e}_d\overset{ind}{\sim}N(0,\sigma^2_{ed})\). We assume that the sampling errors are normally distributed with mean zero and variances \(\sigma^2_{ed}\). Sampling variances \(\sigma^2_{ed}\) usually differ between the different domains, are estimated from the survey, and assumed to be known.
A linking model is used to formulate a model relationship between the parameters of interest \({{\mu}}_d\) and a set of \(p\) area-level auxiliary variables \({\boldsymbol{x}}_d\in \mathbb{R}^{p}\). It is given by
\begin{equation}
{{\mu}}_d = {\boldsymbol{x}}_d^{\top} \boldsymbol{\beta} + u_d,\quad d=1,\ldots,D.
\end{equation}
In each of the \(D\) domains, the parameter of interest, \({{\mu}}_d\), can be written as a linear combination of fixed effects \(\boldsymbol{\beta}\in \mathbb{R}^{p}\) and random effects \(u_d\). For the random effects \(u_d\) we assume distribution \(u_d\overset{iid}{\sim}N(0,\sigma^2_{u})\). This means that we assume all random effect realizations stem from the same distribution with the same variance parameter \(\sigma^2_{u}\).
The sampling and linking model together give the FH model
\begin{equation}
\hat{{\mu}}_d^{Dir} = {\boldsymbol{x}}_d^{\top} \boldsymbol{\beta} + u_d + {{e}}_d,\quad d=1,\ldots,D.
\end{equation}
Sampling errors \(e_d\) and random effects \(u_d\) are assumed to be independent.
In the FH model, we usually assume that the variance parameter \(\sigma^2_{u}\) and the fixed effects \(\boldsymbol{\beta}\) are unknown. They can be estimated via likelihood-based methods such as Maximum Likelihood (ML) and Restricted Maximum Likelihood (REML). These methods strongly depend on the distributional assumptions about the sampling errors \(e_d\) and random effects \(u_d\).
By applying linear mixed model (LMM) theory, we can derive Best Linear Unbiased Predictors (BLUPs) under the FH model. Best means that it is the predictor which minimizes the Mean Squared Error (MSE) under the FH model in the class of linear and unbiased predictors. It is given by
\begin{equation}
\hat{{\mu}}_d^{FH} = {\boldsymbol{x}}_d^{\top} \boldsymbol{\beta} + \gamma_d (\hat{{\mu}}_d^{Dir} – {\boldsymbol{x}}_d^{\top} \boldsymbol{\beta}),\quad d=1,\ldots,D
\end{equation}
with shrinkage factor
\begin{equation}
\gamma_d = \frac{\sigma^2_{u}}{\sigma^2_{u} + \sigma^2_{ed}}.
\end{equation}
When we insert the estimates of variance parameter \(\sigma^2_{u}\) and fixed effects \(\boldsymbol{\beta}\) into the BLUP formula, we get the Empirical BLUPs (EBLUPs). If the chosen model fits the available data well, the EBLUPs can be much more precise than the direct estimates. With the mixed model theory, we can also derive formulas for approximating the MSE of the EBLUPs.
For calculating FH models, we want to point out that it is crucial to carefully select the auxiliary information and check whether the resulting model fits the data at hand.
When you would like to dig deeper into mixed model theory, we recommend you to have a look at the book Mixed Models. If you are especially interested in mixed models for small area estimation, we recommend the book A Course on Small Area Estimation and Mixed Models to you.
Creating Example Fay-Herriot Data
We can use the formulas of the FH model from above to generate example data. We choose \(D=100\) domains of interest and set a seed such that our random variable generations are reproducible.
D <- 100 # Number of domains set.seed(656) # Seed for reproducible results
We simulate 2 auxiliary variables. Their values are drawn from a uniform distribution between 10 and 20. We also create object x_with_intc. It is the auxiliary matrix which additionally contains a column of \(\boldsymbol{1}_D\) as the first column to include an intercept in the model.
x <- sapply(1:2, function (i) { runif (n = D, min = 10, max = 20) }) x_with_intc <- cbind(1, x) # Auxiliary matrix with intercept as first column
We have to choose values for the variance parameters and the fixed effects.
beta <- c(1, 2, 3) sigma2_ed <- rep(2, times = D) sigma2_u <- 2
We choose fixed effects vector \(\boldsymbol{\beta}=(1,2,3)\), the variances of the sampling errors \(\sigma^2_{ed}=2\), \(d=1,\ldots,D\), and the variance of the random effects \(\sigma^2_{u}=2\).
With the above definition, we have all information for drawing the sampling errors \(e_d\) and random effects \(u_d\) from the normal distribution as specified above.
e_d <- sapply(1:D, function (d) { rnorm(1, mean = 0, sd = sqrt(sigma2_ed[d])) # Draw sampling errors }) u_d <- rnorm(D, mean = 0, sd = sqrt(sigma2_u)) # Draw random effects
The vectors of the parameters of interest \(\mu_d\) and their direct estimates \(\hat{{\mu}}_d^{Dir}\) are then calculated as follows.
mu <- x_with_intc %*% beta + u_d # True parameters of interest mu_dir <- mu + e_d # Direct estimates
This is all the data we need to calculate the FH model.
Calculating the Fay-Herriot Model
We are lucky: We do not need to program the functions of the FH model ourselves. Instead, we can use the R package sae.
if (!require('sae', quietly = TRUE)) { install.packages('sae') } library('sae') # Load package 'sae'
With the following line, you can take a look at the documentation of the mseFH function in the package.
?mseFHTo calculate the FH model, we need to have the direct estimates for all domains with their associated variances plus domain-specific auxiliary information. With this information, we can calculate a FH model as follows.
FH_model <- mseFH(formula = mu_dir ~ x, vardir = sigma2_ed, method = "REML") # formula: We model the direct estimates mu_dir as the dependent and the # auxiliary information x as the independent variables. # Function mseFH automatically considers a model intercept. # vardir: Vector of the variances of the direct estimates. # method: Method for estimating fixed effects and variance parameters.
As we know how we generated the data, we can take a look at how well we estimated the true parameters.
# Fixed effects beta # True # [1] 1 2 3 FH_model$est$fit$estcoef # Estimated # beta std.error tvalue pvalue # X(Intercept) 1.422366 1.34996661 1.053631 2.920520e-01 # Xx1 1.935065 0.06585376 29.384275 8.722899e-190 # Xx2 3.058331 0.06359813 48.088372 0.000000e+00 # Variance parameter sigma2_u # True # [1] 2 FH_model$est$fit$refvar # Estimated # [1] 1.616604
The estimates are pretty close. With the presented code you can also conduct a Monte Carlo simulation study (with varying seed) to see whether the parameter estimation is unbiased. Try it out!
Are the FH EBLUPs more efficient than the direct estimates? After all, that’s what matters to us. We can calculate the coefficient of variation (CV) for the direct estimates and the FH EBLUPs.
CV_dir <- sqrt(sigma2_ed) / mu_dir * 100 CV_FH <- sqrt(FH_model$mse) / FH_model$est$eblup * 100
We compare the CVs of the direct estimates and FH EBLUPs visually.
if (!require('ggplot2', quietly = TRUE)) { install.packages('ggplot2') } library('ggplot2') # Load package 'ggplot2' # Generate plot data dat_plot <- data.frame("CV" = c(CV_dir, CV_FH), "Estimator" = c(rep("Direct", D), rep("FH", D)), "Domain" = c(1:D, 1:D)) # Plot the data ggplot(data = dat_plot, aes(x = Domain, y = CV, lty = Estimator, color = Estimator)) + geom_point(pch = 19) + geom_line() + theme(legend.position = "bottom") + scale_y_continuous("Coefficient of variation (CV)")
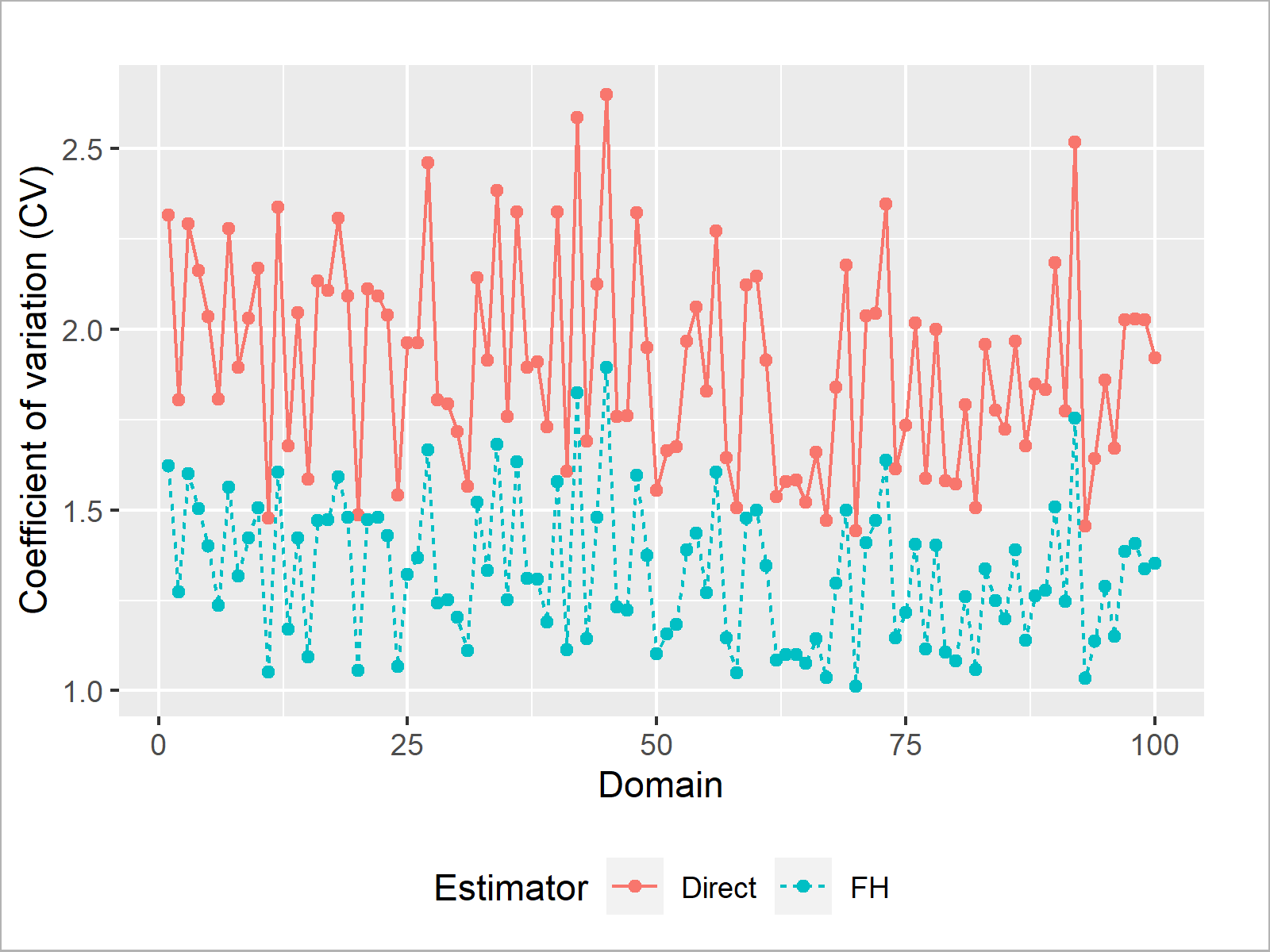
Nice! The FH EBLUPs have a lower CV in all 100 domains. Thus, the FH EBLUPs are more reliable than the direct estimates (assuming that we have verified that the underlying model is also a good fit)!
Video, Further Resources & Summary
For an example of the FH model to actual data, take a look at our article about Small Area Estimation in R.
You want to learn more? For small area estimation in general, we recommend the book Small Area Estimation. There, you can also find some extensions of FH models such as a multivariate version.
If you are interested in more real-world examples of small area estimation, you can have a look at the following YouTube video of the Centre de recherches mathématiques – CRM YouTube channel. Jonathan Wakefield gives a presentation about small area estimation in low and middle-income countries in the video.
Furthermore, have a look at some of the other articles on Statistics Globe:
- Small Area Estimation Techniques
- Small Area Estimation in R
- Battese-Harter-Fuller Small Area Estimator
- Learn R Programming
This post has shown a theoretical introduction into the most famous area-level small area model, the Fay-Herriot model. We further showed how to generate example data from the model and how to calculate the model in the R programming language. If you have further questions, please leave a comment below.
This page was created in collaboration with Anna-Lena Wölwer. Please have a look at Anna-Lena’s author page to get more information about her academic background and the other articles she has written for Statistics Globe.







