R Error in scan: Line 1 did not have X Elements (3 Examples)
In this R tutorial you’ll learn how to handle the error in scan: line 1 did not have X elements.
Table of contents:
Let’s dig in:
Example 1: Reproduce the Error in scan: line 1 did not have X elements
In this example, I’ll explain how to replicate the error message “scan: line 1 did not have X elements” in R.
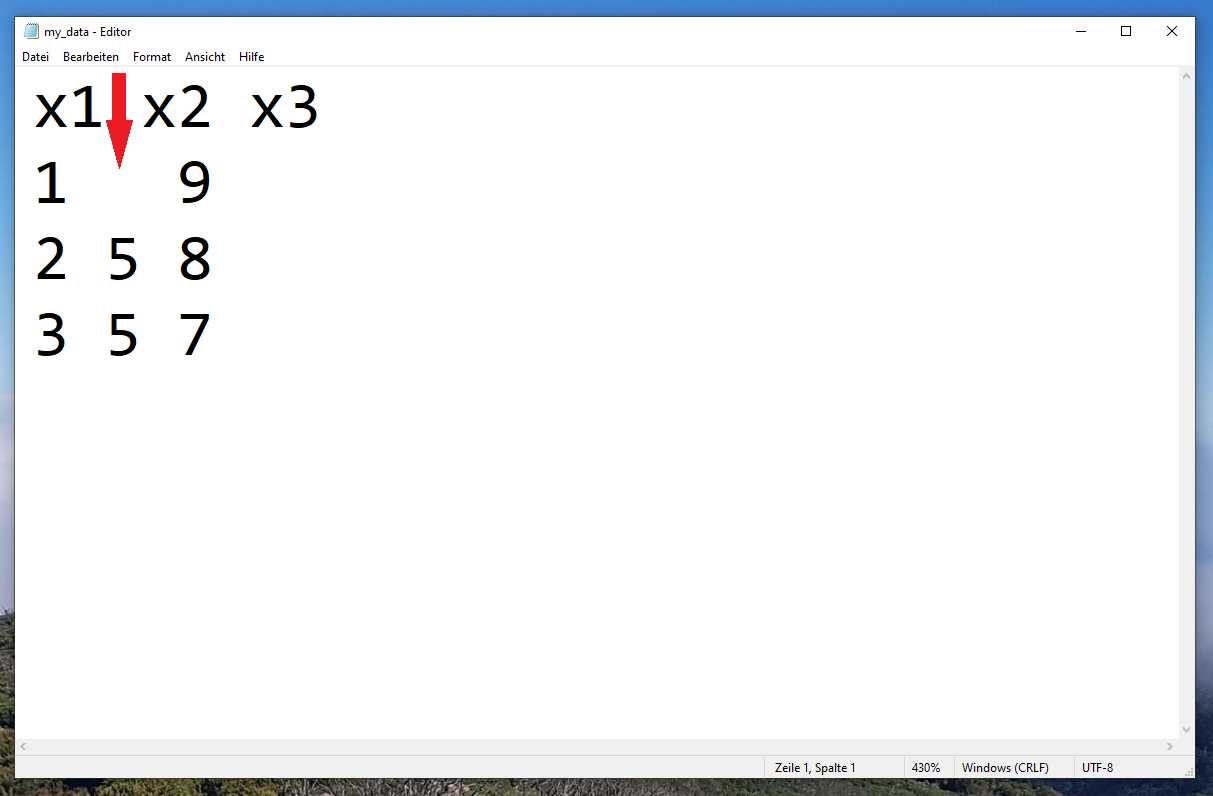
Let’s assume that we want to import the txt-file data set shown in Figure 1:
Then, we could usually use the read.table function as shown below:
read.table("C:/Users/Joach/Desktop/My Folder/my_data.txt", # txt file with empty cells header = TRUE) # Error in scan(file = file, what = what, sep = sep, quote = quote, dec = dec, : # line 1 did not have 3 elements
Unfortunately, the error message “line 1 did not have 3 elements” occurred. The reason for this is that our data is not complete, i.e. some data cells are empty.
Have a look at the first data row (i.e. the row after the header). This row contains only two values, not three values as the other rows.
In the following two examples, I’ll show two different solutions on how to fix this error message. So keep on reading!
Example 2: Fix the Error by Formatting the txt File
In this example, I’ll show how to solve the error in scan: line 1 did not have X elements by formatting our txt input file.
Have a look at the following updated txt-file that I have stored in a new txt-file called “my_data_complete.txt”:
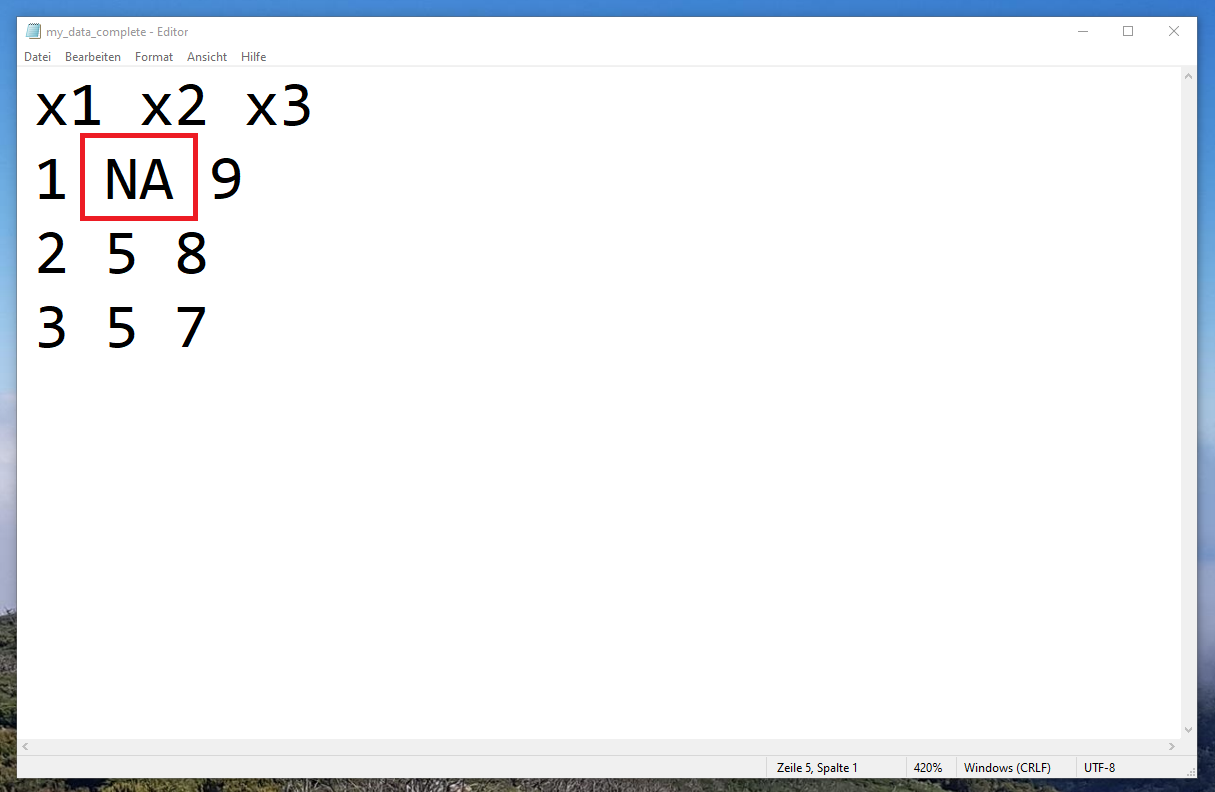
As you can see, I have inserted an NA-value in the second column and the first row of our data.
Let’s import these data into R:
read.table("C:/Users/Joach/Desktop/My Folder/my_data_complete.txt", # Properly formatted txt file header = TRUE) # x1 x2 x3 # 1 1 NA 9 # 2 2 5 8 # 3 3 5 7
Works fine! The data cell in the second variable and the first row is set to a missing value (i.e. NA).
Example 3: Fix the Error Using the fill Argument
The read.table function provides an even simpler solution for importing txt-data with empty cells into R.
We can simply specify the fill argument to be equal to TRUE:
read.table("C:/Users/Joach/Desktop/My Folder/my_data.txt", # Using fill argument header = TRUE, fill = TRUE) # x1 x2 x3 # 1 1 9 NA # 2 2 5 8 # 3 3 5 7
The previous R coded also didn’t lead to any errors. However, please note that the NA value was inserted at the last position of the first row (i.e. the third column). Make sure to check whether this makes sense in your specific data situation!
Video & Further Resources
Do you need further info on the examples of this article? Then you could have a look at the following video of my YouTube channel. I’m explaining the R programming code of this article in the video:
In addition, you might read the other tutorials of this website. I have released numerous articles already.
- scan Function in R
- readLines, n.readLines & readline in R
- Fixing Warning & Error Messages in R
- R Programming Tutorials
In summary: On this page, I showed how to solve problems caused by the error in scan: line 1 did not have X elements in R programming. In case you have additional comments or questions, tell me about it in the comments.







14 Comments. Leave new
Not helpful whatsoever
Clementine, if you provide some more details I might be able to help 😉
Thanks for the valuable examples. but What if you didnt have to insert NA in the data before reading into R especially if data is large and cant be opened except to edit within R?
Hi Elsie,
Thank you for the comment! Have you tried to use the code of Example 3?
Regards
Joachim
fill=TRUE does not work in my case as it shifts the data and misaligns the values to the columns. What is the solution within R to add NAs to empty cells usin read.table? Thanks!
Hey Sanjana,
Excuse me for the delayed response, I took a few days off. Have you already found a solution for this?
Regards,
Joachim
Thanks man. fill=TRUE worked for me.
Awesome, nice to hear that it helped. Thanks for the comment Saad! 🙂
thanks this was helpful
That’s great to hear, thanks for the kind comment Mbwana!
It doesn’t work for me…
Hello,
What is the exact problem in your case? Maybe you consider uploading the data in another format, if possible?
Regards,
Cansu
What if there are no gaps in the table (when viewed in Excel or TextEdit) and it still gives the error message?
Hello Gena,
The error can still occur for other reasons if you’re confident that there are no blanks or extra spaces in your data. Some of the common causes include:
Mismatched delimiters: Make sure the delimiter used in your data file is consistent and matches the one specified in the ‘sep’ parameter of the ‘read.table’, ‘read.csv’, or ‘read.delim’ function. If the delimiters are inconsistent, R might not read the correct number of elements in each row.
Incorrect file encoding: If your data file is not encoded in a format supported by R, it might have trouble reading it correctly. Check the encoding of your file and, if necessary, convert it to a supported format (e.g., UTF-8) before loading it into R.
Missing or extra columns: Double-check that all rows in your data have the same number of columns. If there’s an extra or missing column in any row, it can cause the error. To avoid this, you can use the ‘colClasses’ parameter in the ‘read.table’ function to specify the expected data types for each column.
Embedded quotes or escape characters: Your data might contain quotes (single or double) or escape characters (e.g., ”) within the data values. These characters can cause R to misinterpret the structure of your data. You can resolve this issue by specifying the correct ‘quote’ and ‘escape’ parameters in the ‘read.table’ function.
Comment lines: If your data file contains comment lines starting with a specific character (e.g., ‘#’), specify the ‘comment.char’ parameter in the ‘read.table’ function to ignore these lines.
See the documentation to see how to specify the arguments/parameters mentioned in the suggestions.
Regards,
Cansu