Missing Value Imputation (Statistics) – How To Impute Incomplete Data
It’s an issue every data user knows:
Missing data occur in almost every data set and can lead to serious problems such as biased estimates or less efficiency due to a smaller data set.
To reduce these issues, missing data can be replaced with new values by applying imputation methods.
Missing data imputation is a statistical method that replaces missing data points with substituted values.
In the following step by step guide, I will show you how to:
- Apply missing data imputation
- Assess and report your imputed values
- Find the best imputation method for your data
But before we can dive into that, we have to answer the question…
Why Do We Need Missing Value Imputation?
The default method for handling missing data in R is listwise deletion, i.e. deleting all rows with a missing value in one or more observations.
That approach is easy to understand and to apply, so why should we bother ourselves with more complicated stuff?
Well, it’s as always:
Because we can improve the quality of our data analysis!
The impact of missing values on our data analysis depends on the response mechanism of our data (find more information on response mechanisms here).
Table 1 shows a comparison of listwise deletion (the default method in R) and missing data imputation.
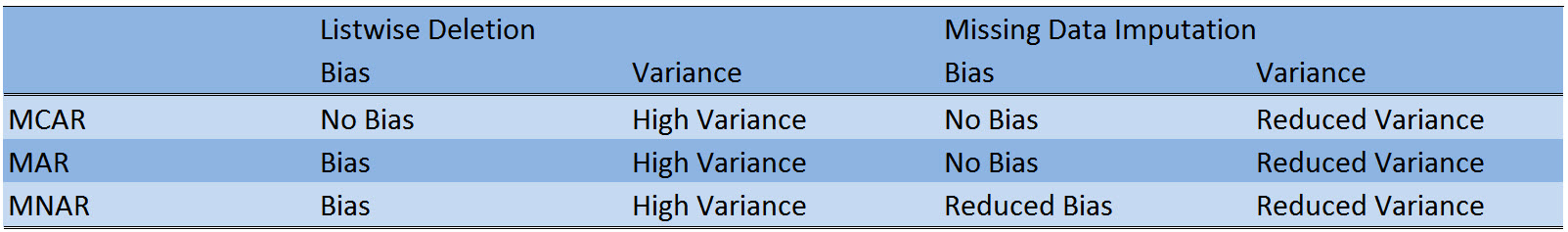
Table 1: Crosstabulation of bias, variance, and the three response mechanisms Missing Completely At Random (MCAR), Missing At Random (MAR), and Missing Not At Random (MNAR).
Table 1 illustrates two major advantages of missing data imputation over listwise deletion:
- The variance of analyses based on imputed data is usually lower, since missing data imputation does not reduce your sample size.
- Depending on the response mechanism, missing data imputation outperforms listwise deletion in terms of bias.
To make it short: Missing data imputation almost always improves the quality of our data!
Therefore we should definitely replace missing values by imputation.
But how does it work? That’s exactly what I’m going to show you now!
Step 1) Apply Missing Data Imputation in R
Missing data imputation methods are nowadays implemented in almost all statistical software.
Below, I will show an example for the software RStudio.
However, you could apply imputation methods based on many other software such as SPSS, Stata or SAS.
The example data I will use is a data set about air quality. In R, the data is already built in and can be loaded as follows:
# Load data data(airquality)
By inspecting the data structure, we can see that there are six variables (Ozone, Solar.R, Wind, Temp, Month, and Day) and 153 observations included in the data. The variables Ozone and Solar.R have 37 and 7 missing values respectively (indicated by NA).
# Data summaries head(airquality)
If we would base our analysis on listwise deletion, our sample size would be reduced to 111 observations.
That’s a loss of 27.5%!
# Check for number of complete cases sum(complete.cases(airquality)) # [1] 111
Such a heavy decrease of our sample size would clearly result in less accuracy and – very likely – also in biased estimates.
Fortunately, with missing data imputation we can do better!
Impute Missing Values in R
A powerful package for imputation in R is called “mice” – multivariate imputations by chained equations (van Buuren, 2017).
The mice package includes numerous missing value imputation methods and features for advanced users.
At the same time, however, it comes with awesome default specifications and is therefore very easy to apply for beginners.
Start by installing and loading the package.
# Install and load the R package mice install.packages("mice") library("mice")
Then, impute missing values with the following code.
# Impute missing data imp <- mice(airquality, m = 1)
After the missing value imputation, we can simply store our imputed data in a new and fully completed data set.
# Store imputed data as new data frame airquality_imputed <- complete(imp)
If you check the structure of our imputed data, you will see that there are no missings left. The imputation process is finished.
# Data summaries of imputed data head(airquality_imputed) summary(airquality_imputed)
That was not so complicated, right?
The reason for that are the predefined default specifications of the mice function.
So let’s have a closer look what actually happened during the imputation process:
m: The argument m was the only specification that I used within the mice function. m = 1 results in a single imputation. Usually, it is preferable to impute your data multiple times, but for the sake of simplicity I used a single imputation in the present example. However, if you want to perform multiple imputation, you can either increase m to the number of multiple imputations you like. Or you can just delete m = 1 from the imputation function for the default specification of five imputed data sets.
method: With the method argument you can select a different imputation method for each of your variables. Mice uses predictive mean matching for numerical variables and multinomial logistic regression imputation for categorical data.
predictorMatrix: Mice automatically uses all available variables as imputation model. In our case, the variables Solar.R, Wind, Temp, Month, and Day were used to impute Ozone and Ozone, Wind, Temp, Month, and Day were used to impute Solar.R. Imputation models can be specified with the argument predictorMatrix, but it often makes sense to use as many variables as possible. Organizational variables such as ID columns can also be dropped using the predictorMatrix argument.
maxit: Imputation was conducted via multivariate imputation by chained equations (Azur et al., 2011). That is another awesome feature of the R package mice. Missing values are repeatedly replaced and deleted, until the imputation algorithm iteratively converges to an optimal value. The mice function repeats the replacement and deletion steps five times by default. With the argument maxit this number can be changed manually.
There are many other arguments that can be specified by the user.
The arguments above, however, are the most important ones.
For further information you can check out the R documentation of mice.
Step 2) Assess and Report Your Imputed Values
To be announced:
- Evaluation of imputed values.
- How to report missing data treatment in scientific publications.
Step 3) Find the Best Imputation Method for Your Data
Imputed values, i.e. values that replace missing data, are created by the applied imputation method.
Researchers developed many different imputation methods during the last decades, including very simple imputation methods (e.g. mean imputation) and more sophisticated approaches (e.g. multiple imputation).
Different methods can lead to very different imputed values.
Hence, it is worth to spend some time for the selection of an appropriate imputation method for your data.
That said, it can be difficult to identify the most appropriate method for your specific database.
The following list gives you an overview about the most commonly used methods for missing data imputation.
Click on the picture in order to get more information about the pros and cons of the different imputation methods.
Articles about the following imputation methods will be announced soon:
- Mean imputation
- Mode imputation for categorical variables
- Replace missing values with 0
- Hot deck nearest neighbor imputation (with and without donor limit)
- Regression imputation (deterministic vs. stochastic)
- Multinomial logistic regression imputation
- Predictive mean matching
Single vs. Multiple Imputation – An Explanation of the Main Concepts
When it comes to data imputation, the decision for either single or multiple imputation is essential.
In single imputation, missing values are imputed just once, leading to one final data set that can be used in the following data analysis.
However, if single imputation is not considered properly in later data analysis (e.g. by applying sophisticated variance estimations), the width of our confidence intervals will be underestimated (Kim, 2011).
By imputing incomplete data several times, the uncertainty of the imputation is taken into account, leading to more precise confidence intervals.
In the following video you can learn more about the advantages of multiple imputation.
The speaker Elaine Eisenbeisz explains the basic concepts of multiple imputation such as Rubins Rules, Pooling of imputed data, and the impact of the response mechanism on imputed values.
She also shows a practical example of multiple imputation with the statistical software SPSS.
References
Melissa J. Azur, Elizabeth A. Stuart, Constantine Frangakis, and Philip J. Leaf (2011). Multiple Imputation by Chained Equations: What is it and how does it work? International Journal of Methods in Psychiatric Research. 20(1): 40–49.
Kim, J.-K. (2001). Variance Estimation After Imputation. Survey Methodology. 27(1):75-83.
van Buuren, S. (2017). Package mice. https://cran.r-project.org/web/packages/mice/mice.pdf
Appendix
The following R code creates the header graphic of this page. The code is based on a graphic of Gaston Sanchez.
The graphic shows the name of this page – Statistical Programming. Some letters are missing.
# Generate X and Y vectors nx <- 100 ny <- 200 x <- sample(x = 1:nx, size = 100, replace = TRUE) y <- seq(- 1, - ny, length = 200) y <- y[y %% 5 == 1] # Set background color op <- par(bg = "#353436", mar = c(0, 0.2, 0, 0.2)) # Create plot plot(1:nx, seq(- 1, - nx), type = "n", xlim = c(1, nx), ylim = c(- 220, 50)) for (i in seq_along(x)) { aux <- sample(1:length(y), 1) # Range of blue colors plot_col <- colorRampPalette(colors=c("#42baff", "#1487ca")) plot_col <- plot_col(20) plot_col <- sample(plot_col) # Letters for "Statistical Programming" plot_let <- c("s", "t", "a", "t", "i", "s", "t", "i", "c", "a", "l", " ", "p", "r", "o", "g", "r", "a", "m", "m", "i", "n", "g", " ") plot_let[rbinom(length(plot_let), 1, 0.35) == 1] <- " " # Plot of loop run i points(rep(x[i], aux), # X values y[1:aux], # Y Values pch = plot_let, # Letters col = plot_col, # Colors cex = runif(aux, 0.75, 1.5)) # Size of letters }







6 Comments. Leave new
Hallo Joachim noch mal :))) I have a small question: I am using this code: data_imp <- mice(Df1)
to impute my dataframe..just as simple as possible, and I knew that mice package picks up automatically the method that fits each variables and it was working so good. I added three categorical variables more "2 categories each" but I got this warning message : ""glm.fit: algorithm did not converge"". I had already before categorical variables but more than 2 categories. I tried to solve this problem, but I couldn't find the solution…I tried to encode my categorical variables but did not help. I tried to create a dataset only from these 3 categorical variables and I did an imputation to this dataset and it works normally.I have only this problem when I do the imputation for the main dataset including these 3 categorical variables all together 🙁 It would be great if you have an idea how to encounter this problem. Thank you so much :)) ….
Hey Mahmoud 🙂
Are those dummy variables predicting each other perfectly? In this case, you might drop one of them. Have a look at this tutorial for more details.
Regards,
Joachim
Thank you sooooooo much..In fact I believe they do so. they are 2 categorical variables that each one indicates the social background of the father and mother, if they are native or from immigrant background. high probably they reflect each other since it is likely that both parents either Native or both non native. I will check your video now. Thank you soooooo much. It took me almost 3 days and lots of research to know the problem behind it..But is there a way to combine both categorical variable in one variable instead of dropping one of them?
Thank you 🙂
Glad it helps!
You may use the predictorMatrix argument of the mice function to drop one of those columns from the imputation model.
Regards,
Joachim
Hello Joachim,
Thank you for your efforts! I have a question: How can I use the weights in the imputation process in case of multilevel data ? Can I easily add the argument of weights and assign the weights variable to the argument in the mice function ?
exemple :
mice(data, method = “pmm”, weight = “weights”)
Also, is it good to add the weights variable? Thank you again.
Hello Ayoub,
I don’t know it by heart. I worked on imputations in another programming language back in the day. However, I did some search. Based on the documentation of the mice package, it looks like you can do it through the weights arguments. I hope it helps. I also see that you want to use the predicted mean matching method. Here is the tutorial written by Joachim about the topic. It can also provide some solutions, hopefully.
Regards,
Cansu